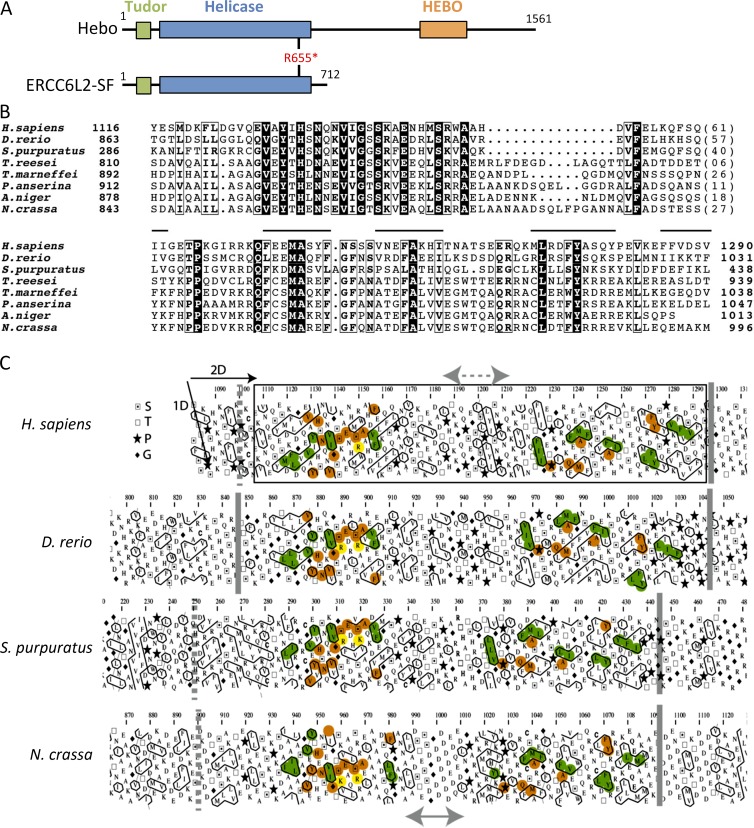
Figure 6.
The HEBO domain. (A) Schematic representation of the Hebo and ERCC6L2-SF domain architecture. The asterisk indicates the stop codon at R655 in the patient. (B) Multiple alignment of the HEBO domain sequence of Hebo proteins from representative species. Identical amino acids are represented as white on a black background, and amino acid similarities are boxed. Lines indicate the position of hydrophobic clusters, which are conserved within the whole family (Table S1). GenBank/NCBI RefSeq accession nos. are NP_064592.2 (Homo sapiens), AAH91795.1 (Danio rerio), XP_011663528 (Strongylocentrotus purpuratus), XP_006968442.1 (Trichoderma reesei), XP_002151909.1 (Talaromyces marneffei), CDP26308.1 (Podospora anserina), XP_001391908.1 (Aspergillus niger), and CAD37001.1 (Neurospora crassa). (C) Hydrophobic cluster analysis of the C-terminal region of ERCC6L6 reveals a globular domain (boxed), which is conserved in the ERCC6L2 orthologues. The sequences are shown on duplicated α-helical nets, on which hydrophobic amino acids are contoured and form clusters, which mainly correspond to the regular secondary structures forming globular domains. Conservation of hydrophobic clusters is indicated in green, whereas sequence identities are reported in orange. A more variable region (arrows) is highlighted in the middle of the domain, likely corresponding either to a large loop or to a linker separating two distinct subdomains. S, T, P, and G stand for serine, threonine, proline, and glycine, respectively. Orange and yellow colorings are used to designate identical and similar amino acids, respectively.