Abstract
The role of exotic plants in regulating soil microbial community structure and activity following invasion chronosequence remains unclear. We investigated soil microbial community structure and microbial respiration following Spartina alterniflora invasion in a chronosequence of 6-, 10-, 17-, and 20-year-old by comparing with bare flat in a coastal wetland of China. S. alterniflora invasion significantly increased soil moisture and salinity, the concentrations of soil water-soluble organic carbon and microbial biomass carbon (MBC), the quantities of total and various types of phospholipid fatty acids (PLFAs), the fungal:bacterial PLFAs ratio and cumulative microbial respiration compared with bare flat. The highest MBC, gram-negative bacterial and saturated straight-chain PLFAs were found in 10-year-old S. alterniflora soil, while the greatest total PLFAs, bacterial and gram-positive bacterial PLFAs were found in 10- and 17-year-old S. alterniflora soils. The monounsaturated:branched PLFAs ratio declined, and cumulative microbial respiration on a per-unit-PLFAs increased following S. alterniflora invasion in the chronosequence. Our results suggest that S. alterniflora invasion significantly increased the biomass of soil various microbial groups and microbial respiration compared to bare flat soil by increasing soil available substrate, and modifying soil physiochemical properties. Soil microbial community reached the most enriched condition in the 10-year-old S. alterniflora community.
Plant invasion, one component of anthropogenic-induced global change, has caused severe biological impacts on native ecosystems and great economic costs1 by changing the composition of species and the ecosystems’ structure2, processes and functioning3,4. Alterations in plant community structure may affect composition of soil microbial community and functioning by altering the quality and quantity of litter input and by modifying soil physical, chemical and biological environment5. Numerous studies have reported that plant invasion can alter the composition of the soil microbial community6,7,8, stimulate or inhibit microbial activity9,10, and change many important nutrient cycling processes and pools4,11. Nevertheless, our understanding of soil microbial community structure and activity as affected by plant invasion is still limited, particularly for different plant invasion chronosequences.
Plant invasion can influence soil microbial community structure and activity by altering the quantity and/or quality of litter entering the soil11,12. Previous studies have found that plant invasion can change aboveground (leaf litter) and belowground (root litter and exudates) inputs13,14. Elgersma et al.8 have reported that the alterations in the soil microbial community are mainly driven by belowground processes (e.g., belowground inputs) rather than aboveground litter inputs8. Plant invasion also shifts the resources available to soil microorganisms and further changes the soil microbial community15. Many invasive plants are considered to be more decay resistant owing to their higher levels of lignins, tannins, and other secondary compounds12, and lower quality of invasive plant materials (higher carbon (C)/nitrogen (N) ratio of litter and/or root)14. These invasive plants are ultimately the cause of higher soil C:N ratios relative to the native ecosystem16. The soil C:N ratio is a primary driver for the alteration in the soil microbial community structure17. The fungal biomass is highly associated with soil C:N ratio, whereas the bacterial biomass is negatively correlated with soil C:N ratio17,18. The response of the soil microbial community structure and activity to plant invasion exhibits high variation, probably as a result of the diverse changes of soil C sources and nutrient availability following invasion of distinct plant species8,10. Alteration in soil substrates during different periods (e.g., short-, mid-, and long-term) of plant invasion can also affect soil microbial community structure and activity19.
Plant invasion can modify the soil’s physical and chemical properties, such as soil moisture16,20, pH21, and salinity16,20, and further affect soil microbial community structure and activity18,22. Soil moisture is a decisive factor of C and N availability and plays a vital role in soil microbial community structure and activity23. The changes in soil moisture can cause alterations in the physiology and growth of some specific soil microbial groups24. Soil pH appears to be the main driving factor for the distribution of soil microorganisms, a higher soil pH would promote gram-negative (gram−) bacteria growth and reduce gram-positive (gram+) bacteria biomass25,26,27. Soil salinity has been considered as one of the important factors for affecting microbial community structure and activity, and high salinity can decrease soil osmotic potential, further affect microbial community composition, decrease microbial biomass and activity28. Thus, the changes in the soil substrates and physicochemical properties altogether affect the soil microbial community structure and activity following plant invasion in a chronosequence.
Spartina alterniflora is a perennial C4 grass plant that is native to North America. It has been introduced to China since 1979 for coastal erosion control and sediment stabilization29,30. S. alterniflora invasion in the coastal zone of China has expanded over the past 30 years, from Tianjin in the north to Beihai in the south, by occupying bare flat and/or by replacing native C3 plants (e.g., Suaeda salsa and Phragmites australis), and become one of the dominant plants in China’s coastal wetland16,20. Previous studies have reported that S. alterniflora has a longer growing season, a higher leaf area index and net photosynthetic rate, and a greater net primary production compared with the native plants, Scirpus mariqueter and P. australis31. Furthermore, S. alterniflora invasion significantly alters soil physicochemical properties16, soil organic C and N sequestration14,29, and emissions of greenhouse gases in the coastal wetland of eastern China20. However, little is known about the changes in the soil microbial community structure and activity in chronosequences following S. alterniflora invasion. We hypothesized that S. alterniflora invasion would alter soil microbial community structure and activity by changing soil C availability and physiochemical properties. To test this hypothesis, we determined soil phospholipid fatty acids (PLFAs) to analyze the soil microbial community structure, and determined cumulative microbial respiration, microbial respiration on a per-unit-PLFAs basis, and the respiration quotient (qCO2) after 30-days of incubation at 25 °C and 35 °C to analyze the soil microbial activity. We measured soil moisture, pH, salinity, soil organic C (SOC), soil organic N (SON), water-soluble organic carbon (WSOC), microbial biomass C (MBC), microbial biomass N (MBN), the MBC:MBN ratio, temperature sensitivity (Q10) of microbial respiration, and the aboveground and root biomass in invasive 6-, 10-, 17-, and 20-year-old S. alterniflora communities and compared these findings with those from a bare flat in a coastal wetland of China.
Results
Soil and plant properties
Soil moisture, salinity, WSOC, SOC, and SON in S. alterniflora soils were significantly higher than those in bare flat soil (Table 1). Soil moisture was the highest in 17- and 20-year-old S. alterniflora soils followed by 6- and 10-year-old S. alterniflora soils (Table 1). The pH in S. alterniflora soils were significantly lower than that in bare flat soil with the lowest pH in 6- and 17-year-old S. alterniflora soils (Table 1). The highest salinity and the lowest WSOC were found in 20-year-old S. alterniflora soil, while the greatest SOC concentration was found in 17-year-old S. alterniflora soil (Table 1). Aboveground biomass was the highest in the 17-year-old S. alterniflora community, followed by 20-, 10-, and 6-year-old S. alterniflora communities (Table 1). However, SON concentration and root biomass did not significantly change across the S. alterniflora invasion chronosequence (Table 1).
Table 1. Soil (0–30 cm depth) and plant properties (mean ± SE, n = 9) following S. alterniflora invasion in a coastal wetland of China.
| Moisture (%) | pH | Salinity (%) | SOC (g kg−1) | WSOC (mg kg−1) | SON (g kg−1) | Aboveground biomass (g m−2) | Root biomass (g m−2) | |
|---|---|---|---|---|---|---|---|---|
| Bare flat | 19.67 ± 0.37c | 8.87 ± 0.02a | 0.66 ± 0.04c | 0.95 ± 0.02c | 28.33 ± 0.34d | 0.219 ± 0.035b | – | – |
| S. alterniflora | ||||||||
| 6 years | 45.45 ± 0.86b | 8.48 ± 0.04c | 1.82 ± 0.20b | 10.07 ± 1.01b | 55.62 ± 0.38b | 1.019 ± 0.168a | 1777 ± 137c | 5530 ± 468a |
| 10 years | 46.57 ± 0.47b | 8.59 ± 0.03b | 1.85 ± 0.17b | 10.25 ± 1.92b | 61.92 ± 1.74a | 1.058 ± 0.245a | 1845 ± 138c | 5808 ± 601a |
| 17 years | 52.54 ± 0.39a | 8.46 ± 0.03c | 1.78 ± 0.10b | 15.56 ± 0.50a | 62.14 ± 1.01a | 1.357 ± 0.039a | 3009 ± 175a | 5291 ± 269a |
| 20 years | 51.17 ± 0.33a | 8.54 ± 0.03bc | 2.23 ± 0.07a | 11.92 ± 0.64ab | 47.74 ± 1.19c | 1.135 ± 0.039a | 2330 ± 116b | 5435 ± 707a |
| Source of variation | ||||||||
| Invasion time | *** | *** | *** | *** | ** | ** | *** | n.s. |
Different letters indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence. **P < 0.01; ***P < 0.001; n.s.: not significant; SOC: soil organic carbon; WSOC: soil water-soluble organic carbon; SON: soil organic nitrogen.
Soil microbial biomass and structural diversity
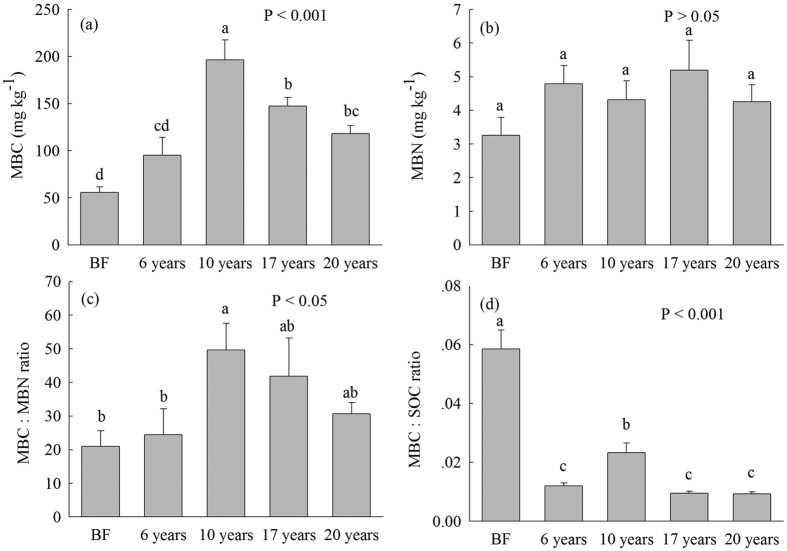
The highest MBC concentration was found in 10-year-old S. alterniflora soil, followed by 17-, 20-, 6-year-old S. alterniflora and bare flat soils (Fig. 1a). In contrast, MBN did not significantly vary in the S. alterniflora invasion chronosequence (Fig. 1b). The MBC:MBN ratio in 10-year-old S. alterniflora soil was significantly higher than that in 6-year-old S. alterniflora and bare flat soils (Fig. 1c). The MBC:SOC ratio in bare flat soil was significantly higher than that in S. alterniflora soils, and the MBC:SOC ratio in 10-year-old S. alterniflora soil was significantly higher than that in 6-, 17-, 20-year-old S. alterniflora soils (Fig. 1d). MBC concentration was strongly associated with soil and plant properties except soil pH (Table 2).
Figure 1.
(a) Soil microbial biomass carbon (MBC), (b) Soil microbial biomass nitrogen (MBN), (c) MBC:MBN ratio and (d) MBC:SOC ratio (mean ± SE, n = 9) in bare flat (BF) and different invasion times (6, 10, 17 and 20 years) of S. alterniflora soils (0–30 cm depth). Different lower case letters over the bars indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence.
Table 2. Pearson correlation coefficients between soil microbial indexes and the soil and plant properties across the communities.
| Moisture | pH | Salinity | SOC | WSOC | SON | Aboveground biomass | Root biomass | |
|---|---|---|---|---|---|---|---|---|
| MBC | 0.563** | −0.469** | 0.479** | 0.483** | 0.747** | 0.480** | 0.498** | 0.569** |
| MBN | 0.263 | −0.343* | 0.238 | 0.235 | 0.308* | 0.199 | 0.326* | 0.336* |
| MBC:MBN | 0.281 | −0.144 | 0.252 | 0.333* | 0.381** | 0.344* | 0.192 | 0.215 |
| Total PLFAs | 0.754** | −0.720** | 0.734** | 0.676** | 0.796** | 0.674** | 0.651** | 0.649** |
| Bacterial PLFAs | 0.752** | −0.713** | 0.726** | 0.658** | 0.785** | 0.660** | 0.631** | 0.642** |
| Fungal PLFAs | 0.660** | −0.658** | 0.683** | 0.644** | 0.801** | 0.640** | 0.621** | 0.656** |
| Gram+ bacterial PLFAs | 0.781** | −0.751** | 0.735** | 0.718** | 0.812** | 0.705** | 0.698** | 0.661** |
| Gram− bacterial PLFAs | 0.736** | −0.696** | 0.714** | 0.649** | 0.774** | 0.653** | 0.621** | 0.619** |
| AMF PLFAs | 0.687** | −0.687** | 0.678** | 0.504** | 0.689** | 0.540** | 0.461** | 0.581** |
| Actinomycete PLFAs | 0.680** | −0.664** | 0.615** | 0.798** | 0.789** | 0.747** | 0.791** | 0.614** |
| Monounsaturated PLFAs | 0.711** | −0.679** | 0.706** | 0.583** | 0.737** | 0.600** | 0.548** | 0.600** |
| Branched PLFAs | 0.772** | −0.743** | 0.719** | 0.757** | 0.832** | 0.733** | 0.740** | 0.673** |
| SSC PLFAs | 0.752** | −0.700** | 0.756** | 0.618** | 0.747** | 0.626** | 0.592** | 0.634** |
| CMR at 25 °C | 0.911** | −0.796** | 0.876** | 0.735** | 0.718** | 0.704** | 0.738** | 0.688** |
| CMR at 35 °C | 0.901** | −0.775** | 0.822** | 0.750** | 0.713** | 0.721** | 0.736** | 0.656** |
| Q10 value | 0.025 | −0.025 | −0.123 | 0.160 | 0.103 | 0.180 | 0.083 | 0.008 |
*P < 0.05; **P < 0.01. MBC: microbial biomass carbon; MBN: microbial biomass nitrogen; PLFAs: phospholipid fatty acids; Gram+: gram-positive; Gram−: gram-negative; AMF: arbuscular mycorrhizal fungal; SSC: saturated straight-chain; CMR: cumulative microbial respiration; Q10: temperature sensitivity; See Table 1 for abbreviations.
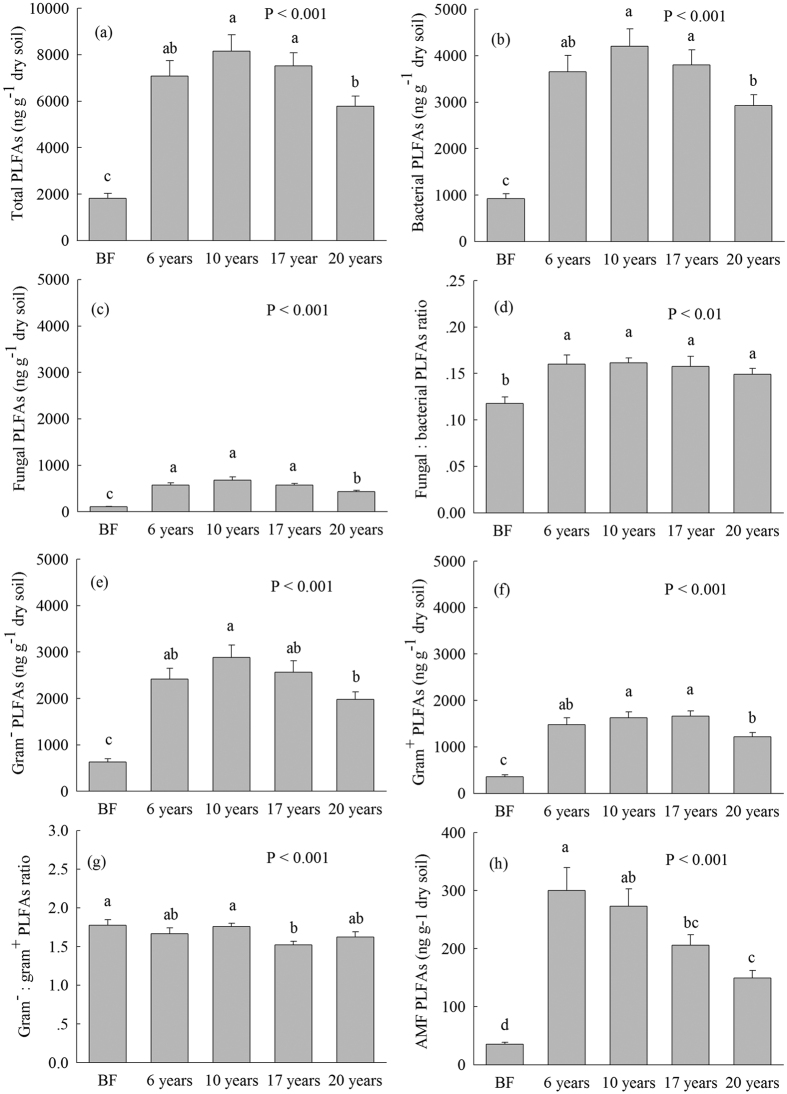
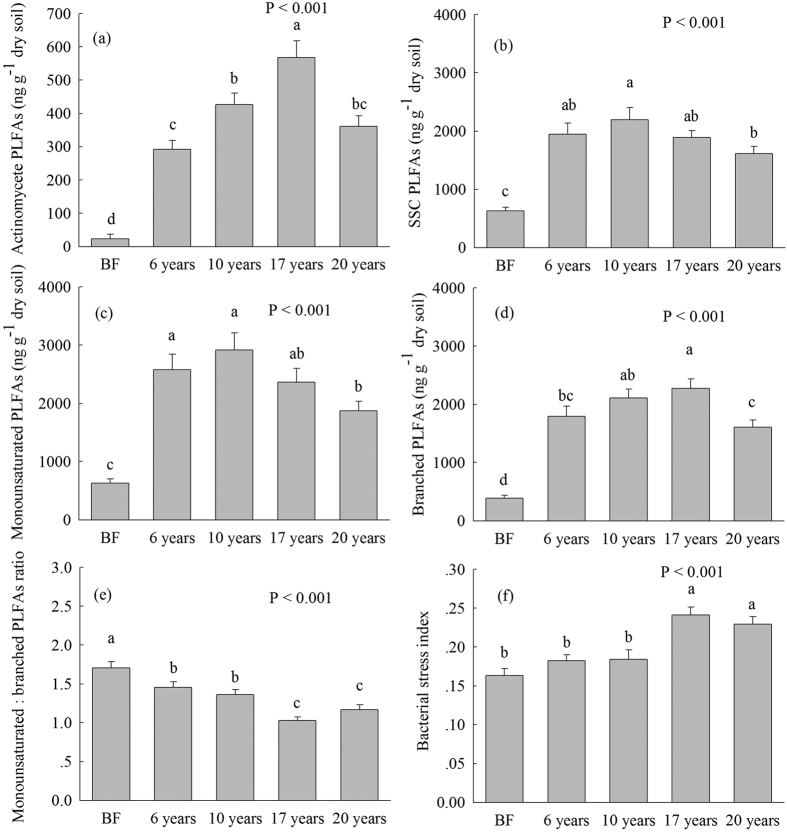
The quantities of the total PLFAs, bacterial, fungal, gram+ bacterial, gram− bacterial, arbuscular mycorrhizal fungi (AMF), actinomycete, monounsaturated, branched, and saturated straight-chain (SSC) PLFAs in S. alterniflora soils were significantly higher than those in bare flat soil (Figs 2 and 3). The quantities of soil total PLFAs, bacterial, and gram+ bacterial PLFAs were the highest at 10 and 17 years, followed by 6 years and 20 years after S. alterniflora invasion (Fig. 2a,b,f). The lowest fungal PLFAs were found in 20-year-old S. alterniflora soil (Fig. 2c). The quantity of AMF PLFAs gradually declined following S. alterniflora invasion in the chronosequence (Fig. 2h). The quantities of actinomycete and branched PLFAs gradually increased from 6 to 17 years after S. alterniflora invasion, but declined in the 20-year-old S. alterniflora soil (Fig. 3a,d). The quantities of gram− bacterial and SSC PLFAs were the greatest in 10-year-old S. alterniflora soil and the lowest in 20-year-old S. alterniflora soil (Figs 2e and 3b). The quantity of monounsaturated PLFAs was the most enriched in 10- and 6-year S. alterniflora soils, and it declined in 17- and 20-year S. alterniflora soils (Fig. 3c).
Figure 2.
(a) The total phospholipid fatty acids (PLFAs), (b) Bacterial PLFAs, (c) Fungal PLFAs concentrations; (d) Fungal:bacterial PLFAs ratio; (e) Gram− PLFAs, (f) Gram+ PLFAs concentrations, (g) Gram−:gram+ PLFAs ratio and (h) The arbuscular mycorrhizal fungal PLFAs (AMF PLFAs) concentration (mean ± SE, n = 9) in bare flat (BF) and different invasion times (6, 10, 17 and 20 years) of S. alterniflora soils (0–30 cm depth). Different lower case letters over the bars indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence.
Figure 3.
(a) Actinomycete phospholipid fatty acids (PLFAs), (b) The saturated straight-chain (SSC) PLFAs, (c) The monounsaturated PLFAs, (d) Branched PLFAs concentrations, (e) Monounsaturated:branched PLFAs ratio and (f) Bacterial stress index (mean ± SE, n = 9) in bare flat (BF) and different invasion times (6, 10, 17 and 20 years) of S. alterniflora soils (0–30 cm depth). Different lower case letters over the bars indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence.
The fungal:bacterial PLFAs ratio in all S. alterniflora soils were considerably higher than that in bare flat soil (Fig. 2d). The lowest gram−:gram+ ratio was found in 17-year-old S. alterniflora soil, and the highest gram−:gram+ ratio was found in 10-year-old S. alterniflora and bare flat soils (Fig. 2g). The monounsaturated:branched PLFAs ratio in 17- and 20-year-old S. alterniflora soils were considerably lower than that in 6- and 10-year-old S. alterniflora soils (Fig. 3e). The bacterial stress index in 17- and 20-year-old S. alterniflora soils were significantly higher than that in 6- and 10-year-old S. alterniflora and bare flat soils (Fig. 3f).
Soil microbial respiration and temperature sensitivity
Cumulative microbial respiration in the 0–30 cm soil layer after 30 days of incubation at 35 °C was significantly greater than soil from the bare flat and S. alterniflora invasion chronosequence incubated at 25 °C (Fig. 4a). Cumulative microbial respiration in over 10-year-old S. alterniflora soils were considerably higher than that in 6-year-old S. alterniflora soil, which was higher compared with bare flat soil (Fig. 4a). Cumulative microbial respiration was not only significantly related to soil moisture, salinity, SOC, WSOC, SON, and aboveground and root biomass (Table 2) but also strongly associated with total and all types of PLFAs (Table 3).
Figure 4.
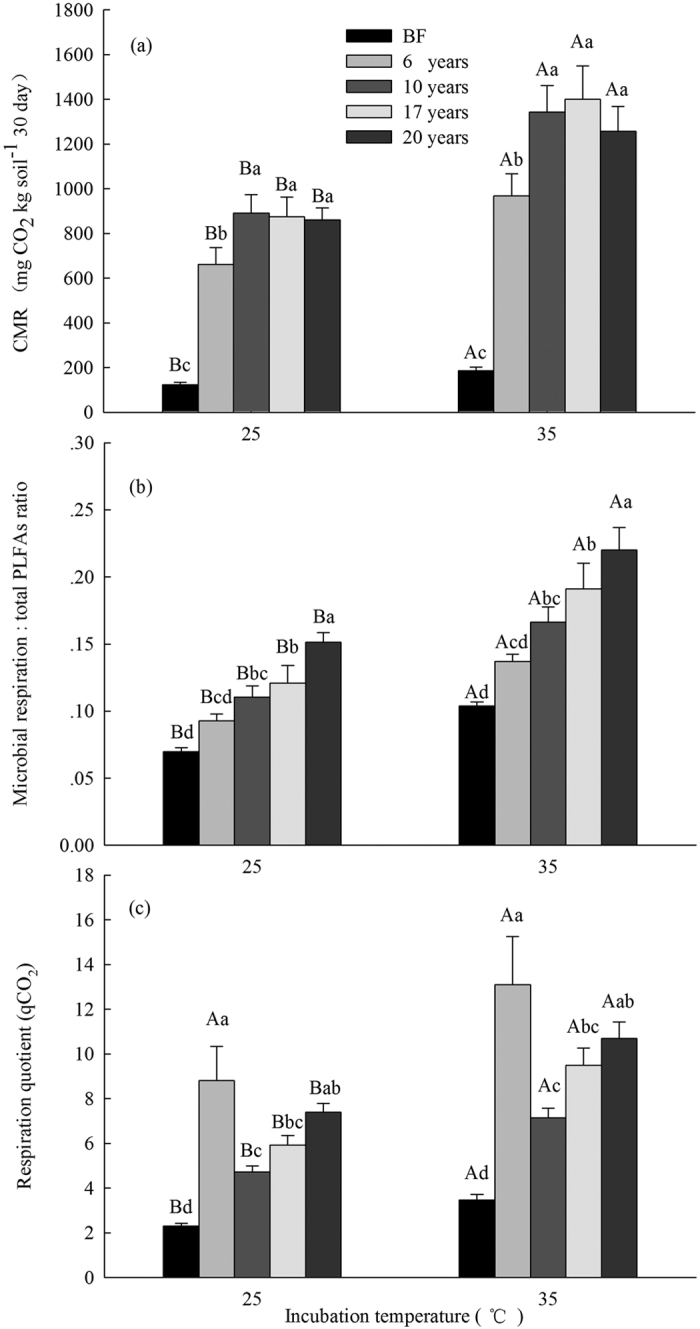
(a) Cumulative microbial respiration (CMR), (b) Microbial respiration on a per-unit-PLFAs basis and (c) Respiration quotient (qCO2) after 30-days incubation under different temperature treatments (25 °C and 35 °C) (mean ± SE, n = 9) in bare flat (BF) and different invasion times (6, 10, 17 and 20 years) of S. alterniflora soils (0–30 cm depth). Different lower case letters over the bars indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence. Different upper case letters over the bars indicate statistically significant differences at α = 0.05 level between incubation temperature.
Table 3. Pearson correlation coefficients between soil microbial respiration and microbial biomass across the communities.
| Total PLFAs | Bacterial PLFAs | Fungal PLFAs | Gram+ bacterial PLFAs | Gram− bacterial PLFAs | AMF PLFAs | Actinomycete PLFAs | Monounsaturated PLFAs | Branched PLFAs | SSC PLFAs | |
|---|---|---|---|---|---|---|---|---|---|---|
| CMR at 25 °C | 0.811** | 0.808** | 0.748** | 0.813** | 0.800** | 0.686** | 0.729** | 0.770** | 0.813** | 0.813** |
| CMR at 35 °C | 0.817** | 0.815** | 0.735** | 0.824** | 0.812** | 0.676** | 0.772** | 0.769** | 0.832** | 0.800** |
| MRP at 25 °C | 0.210 | 0.201 | 0.184 | 0.236 | 0.191 | 0.091 | 0.291 | 0.148 | 0.258 | 0.217 |
| MRP at 35 °C | 0.249 | 0.241 | 0.200 | 0.283 | 0.234 | 0.112 | 0.370* | 0.176 | 0.315* | 0.234 |
| qCO2 at 25 °C | 0.486** | 0.471** | 0.419** | 0.495** | 0.465** | 0.434** | 0.453** | 0.440** | 0.491** | 0.508** |
| qCO2 at 35 °C | 0.510** | 0.498** | 0.425** | 0.527** | 0.494** | 0.444** | 0.511** | 0.456** | 0.531** | 0.515** |
| Q10 value | 0.128 | 0.133 | 0.056 | 0.151 | 0.146 | 0.060 | 0.253 | 0.097 | 0.180 | 0.058 |
*P < 0.05; **P < 0.01. MRP: microbial respiration: total PLFAs ratio; qCO2: respiration quotient; See Table 2 for abbreviations.
Similarly, microbial respiration on a per-unit-PLFAs and qCO2 after 30-days of incubation at 35 °C were significantly higher than in soils from all communities that were incubated at 25 °C, the exception being qCO2 in 6-year-old S. alterniflora soil (Fig. 4b,c). Microbial respiration on a per-unit-PLFAs gradually increased following the S. alterniflora invasion chronosequence (Fig. 4b). The qCO2 in 6- and 20-year-old S. alterniflora soils were significantly higher than that in 10-year-old S. alterniflora and bare flat soils (Fig. 4c). The Q10 value of microbial respiration did not significantly change across bare flat soil and the S. alterniflora invasion chronosequence (Fig. 5). Pearson’s correlation analysis showed that qCO2 was significantly associated with total and all types of PLFAs (Table 3) and that the Q10 value of microbial respiration was not significantly related to soil and plant properties (Table 2), and it was also unrelated to total and all types of PLFAs (Table 3).
Figure 5. Temperature sensitivity (Q10) of microbial respiration after 30-days incubation time at 25 °C and 35 °C (mean ± SE, n = 9) in bare flat (BF) and different invasion times (6, 10, 17 and 20 years) of S. alterniflora soils (0–30 cm depth).
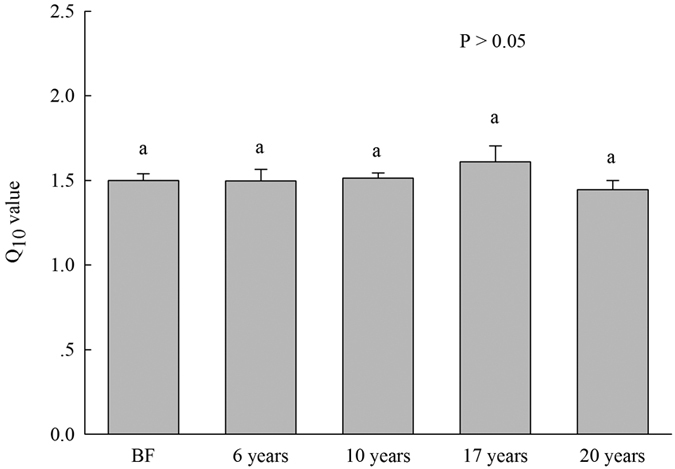
Different lower case letters over the bars indicate statistically significant differences at α = 0.05 level across the S. alterniflora invasion chronosequence.
Controls on soil microbial community
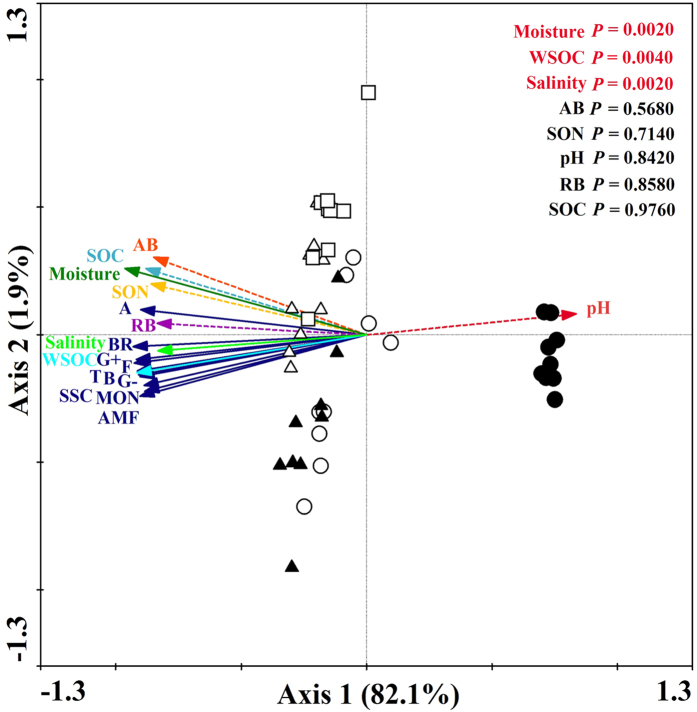
Eight variables of soil and plant properties, including, soil moisture, pH, salinity, SOC, WSOC, SON, aboveground and root biomass, explained 84.1% of the total variability in the PLFAs (Fig. 6). The variations in the PLFAs were strongly correlated with soil moisture (F = 131.16, P = 0.0020), WSOC (F = 8.75, P = 0.0040), and salinity (F = 10.07, P = 0.0020) (Fig. 6). The biggest variation, at 82.1%, was explained by the total variations of the PLFAs in Axis 1, and Axis 2 explained 1.9% of the total variations of the PLFAs (Fig. 6). Meanwhile, Pearson’s correlation analysis showed that the PLFAs were significantly positively correlated with soil moisture, salinity, SOC, WSOC, SON, aboveground and root biomass, but they were negatively associated with soil pH (Table 2).
Figure 6. RDA results of PLFAs in the soil samples and soil and plant properties.
The explanatory variables are showed by different arrows: PLFAs profiles by blue solid arrows: total PLFAs (T); bacterial PLFAs (B); fungal PLFAs (F); gram-positive bacterial PLFAs (G+); gram-negative bacterial PLFAs (G−); arbuscular mycorrhizal fungal PLFAs (AMF); actinomycete PLFAs (A); saturated straight-chain PLFAs (SSC), monounsaturated PLFAs (MON); branched PLFAs (BR); and the variables of soil and plant properties by colored arrow: soil moisture, pH, salinity, soil organic carbon (SOC), soil water-soluble organic carbon (WSOC), soil organic nitrogen (SON), aboveground biomass (AB) and root biomass (RB). Filled circles represent bare flat soil, open circles represent 6-year-old S. alterniflora soil, filled triangles represent 10-year-old S. alterniflora soil, open triangles represent 17-year-old S. alterniflora soil, and open squares represent 20-year-old S. alterniflora soil.
Discussion
Our findings not only added to various evidence that S. alterniflora invasion greatly accelerated soil organic C and N accumulation due to greater biomass input14,29 (Table 1), but also found that S. alterniflora invasion significantly increased MBC concentration and the quantities of the total and all types of PLFAs compared with bare flat soil (Figs 1, 2, 3). Soil C sources are considered as crucial ecological driving factors for microbial community dynamics32. Increased biomass input and the soil substrate following S. alterniflora invasion14,16 (Table 1) possibly enhanced MBC and all types of PLFAs. This speculation was supported by our Pearson’s correlation analysis that MBC and all types of PLFAs were highly associated with SOC, WSOC, SON, and above- and below- ground biomass (Table 2). The aboveground biomass and SOC content progressively increased in 6- to 17-year-old S. alterniflora soils and then fell in soils collected afterwards (Table 1). Interestingly, the highest MBC, gram− bacterial and SSC PLFAs were found in 10-year-old S. alterniflora soil (Figs 1, 2, 3), and the greatest total PLFAs, bacterial and gram+ bacterial PLFAs were found in 10- and 17-year-old S. alterniflora soils (Fig. 2), implying that the soil microbial community reached the most enriched condition in 10-year-old S. alterniflora soil. The WSOC is the most important available substrate and directly provides available C and energy for soil microbial metabolism33,34. Although 17-year-old S. alterniflora soil had a bigger SOC stock compared with 10-year-old S. alterniflora soil (Table 1), there was no significant difference in the quantity of total PLFAs between 10- and 17-year-old S. alterniflora soils (Fig. 2) due to the same level of WSOC in both samples (Table 1). Meanwhile, the decrease in total PLFAs, bacterial, fungal, gram+ bacterial, and branched PLFAs in 20-year-old S. alterniflora soil compared with 10- and 17-year-old S. alterniflora soils (Figs 2 and 3), may be caused by lower levels of readily available substrate (i.e., WSOC; Table 1), which restricted soil microbial growth and metabolism. The RDA and Pearson’s correlation analysis confirmed that the variations in the all types of PLFAs were highly related to WSOC (Table 2, Fig. 6).
The fungal:bacterial PLFAs ratio can be used to reflect the physiological state of the soil microbial community that is particularly involved in SOM accumulation and turnover35, and the ecosystem’s buffering capacity36. In this study, S. alterniflora invasion significantly increased the fungal:bacterial PLFAs ratio compared with bare flat soil (Fig. 2d), indicating a higher C accumulation and self-buffering capacity in S. alterniflora soils. Previous studies reported that fungi have higher C assimilation efficiency relative to bacteria37,38, owing to their stronger ability to decompose plant compounds39,40. Higher C assimilation efficiency in fungi may result in more organic C being converted into more recalcitrant humic materials37. Hence, the increased fungal:bacterial PLFAs ratio in S. alterniflora soil can possibly enhance soil organic C sequestration following S. alterniflora invasion.
We found that the gram−:gram+ PLFAs ratio ranged from 1.52 to 1.78 across S. alterniflora invasion chronosequence (Fig. 2g), suggesting that gram− bacteria dominated in bare flat and S. alterniflora salt marsh and that there were copiotrophic condition in this coastal wetland ecosystem22. Previous studies showed that higher soil pH would increase gram− bacteria and decrease gram+ bacteria25,26,27. 10-year-old S. alterniflora and bare flat soils had a higher pH compared with 17-year-old S. alterniflora soil (Table 1), and this may be one of the reasons that the lowest gram−:gram+ PLFAs ratio was found in 17-year-old S. alterniflora soil (Fig. 2g). This result was consistent with our finding that 17- and 20-year-old S. alterniflora soils had a greater bacterial stress index than 10- and 6-year-old S. alterniflora and bare flat soils (Fig. 3f). Generally, a high bacterial stress index represents a slow rate of growth and long turnover time for gram− bacteria41. Thus, higher bacterial stress index in 17- and 20-year-old S. alterniflora soils indicated that they had slower growth rates and lower turnover rates of the gram− bacteria community relative to 10- and 6-year-old S. alterniflora and bare flat soils (Fig. 3f). Additionally, previous studies have reported that gram− bacteria preferentially utilize fresh plant residual as an available C source, while gram+ bacteria prefer to use older, humified and more microbially processed SOM42,43. Thus, the lowest gram−:gram+ PLFAs ratio and the highest bacterial stress index were found in 17-year-old S. alterniflora soil (Figs 2g and 3f), indirect suggesting increased degree of SOM decomposition and humification in 17-year-old S. alterniflora soil compared with 10-year-old S. alterniflora and bare flat soils.
The monounsaturated and branched PLFAs were generally used to indicate aerobic and anaerobic microorganism biomass, respectively41,44. We found that the highest levels of monounsaturated PLFAs were found in 10- and 6-year-old S. alterniflora soils (Fig. 3c), while the highest levels of branched PLFAs was found in 17-year-old S. alterniflora soil across the invasion chronosequence (Fig. 3d), suggesting that the quantity of aerobic microbes was the highest in the early stage of S. alterniflora invasion, whereas the quantity of anaerobic microbes reached maximum levels at the later stage of S. alterniflora invasion. The ratio of monounsaturated:branched PLFAs continually declined during the invasion chronosequence (Fig. 3e), implying that the percent of anaerobic microbes gradually increased and the percent of aerobic microbes gradually decreased during the invasion chronosequence. This may be highly associated with the gradual increase in soil moisture during the invasion chronosequence (Table 1). Higher soil moisture provides stronger soil anaerobic conditions, which might be more suitable for anaerobic microorganism growth and facilitate SOM accumulation45,46.
Soil microbial respiration was highly dependent on soil temperature, moisture, and C inputs47, and was strongly associated with the quantities of the soil microbes and WSOC concentration34. In this study, cumulative microbial respiration at 25 °C and 35 °C at different invasion times of S. alterniflora soils were significantly higher than that in the bare flat (Fig. 4a), which was highly correlated with total and various types of PLFAs, WSOC, aboveground and root biomass (Tables 2 and 3). Thus, the increased cumulative microbial respiration in S. alterniflora soils may be greatly attributed to higher C inputs, and increase in available substrate (e.g., WSOC) and the microbial biomass (Table 1; Figs 1, 2, 3, 4). We found that cumulative microbial respiration and the microbial respiration:total PLFAs ratio at 35 °C in all communities were significantly higher than that at 25 °C (Fig. 4a,b), primarily because elevated temperature increases soil enzyme activities and further drives SOM decomposition48. Interestingly, the Q10 value of microbial respiration showed no obvious changes during the S. alterniflora invasion chronosequence (Fig. 5), likely because the Q10 of microbial respiration is not influenced by the differences in the microflora34 (Table 3). Although cumulative microbial respiration at 25 °C and 35 °C showed no significant differences between 10-, 17-, and 20-year-old S. alterniflora soils (Fig. 4a), cumulative microbial respiration on a per-unit-PLFAs basis at 25 °C and 35 °C progressively increased following the increase of invasion time (Fig. 4b), suggesting that S. alterniflora invasion may decrease microbial C utilization efficiency and enhance respiration loss in this coastal wetland ecosystem48. Generally, an increase in qCO2 may reflect a decrease in microbial C utilization efficiency and ecosystem stabilization36,49, and a higher MBC:SOC ratio could indicate an increase in microbes use C efficiency16,50. In this study, the lowest qCO2 and the highest MBC:SOC ratio were found in 10-year-old S. alterniflora soil following invasion from 6 to 20 years (Figs 1d and 4c), indicating that 10-year-old S. alterniflora soil had the highest microbial C utilization efficiency and the greatest ecosystem stabilization following invasion from 6 to 20 years. In addition, bare flat soil had higher the MBC:SOC ratio and lower cumulative microbial respiration on a per-unit-PLFAs and qCO2 relative to S. alterniflora soils (Figs 1d and 4b,c), implying that S. alterniflora invasion resulted in low microbial C utilization efficiency compared to bare flat16,50, which may be due to S. alterniflora with lower quality and more recalcitrant substance (e.g., lignin) is difficult to be utilized by microbes14.
In conclusion, this study highlighted the variations of soil microbial community structure and activity following bare flat was converted to S. alterniflora salt marsh in a invasion chronosequence in a coastal wetland of China. Specifically, S. alterniflora invasion greatly increased the total and various types of soil microbial biomass and cumulative microbial respiration but significantly decreased microbial C utilization efficiency compared to bare flat. 10-year-old S. alterniflora community had the most enriched soil microbial community and the highest microbial C utilization efficiency across 6 to 20 years S. alterniflora invasion. Soil microbial biomass decreased after 17 years S. alterniflora invasion. The variations in the microbial community structure and activity may in turn deeply affect SOM accumulation and ecosystem C and N cycling. This study represents a step forward in our understanding of microbial communities as affected by plant invasion, and it provides valuable insights regarding the better understand the influence mechanism of plant invasion on soil organic C pool.
Methods
Site description and sampling
This study was conducted at the core area of the Jiangsu Yancheng Wetland National Nature Reserve, Rare Birds, China (JYWNNRRB) (32°48′47″–34°29′28″N, and 119°53′45″–121°18′12″E). This area is characterized as warm temperate with an average annual temperature of 13.8 °C, average annual precipitation of 1000 mm, and average annual sea water salinity of 3.09%16. JYWNNRRB was designated as an internationally important wetland site (Ramsar) in 2002. S. alterniflora was introduced to the bare flat of the JYWNNRRB in 1983, and it quickly expanded to form large areas of S. alterniflora salt marshes following mudflat aggrading14. The bare flat and S. alterniflora salt marshes are located on the low and middle areas of the intertidal zone with semidiurnal tidal periodicity20. The seaward invasion region of S. alterniflora is a bare flat that had no vegetation prior to S. alterniflora invasion16.
The sampling region, with its different S. alterniflora invasion times, was identified based on analyses of Landsat Thematic Mapper 5 (TM5) satellite images and historical records. This chronosequence from seaward to landward contained the bare flat (BF) and the four S. alterniflora communities that were colonized in 2006 (6 years), 2002 (10 years), 1995 (17 years) and 1992 (20 years). In November 2012, three parallel transects (2-km length and 200-m width) were selected along the chronosequence. Within each transect, five locations were marked from the bare flat to the invasive 6, 10, 17, and 20 years S. alterniflora communities. Three 2 m × 2 m plots were randomly established within each location. Three soil samples (5-cm diameter × 30 cm depth) were collected randomly in each plot. The soil samples from each plot were mixed evenly to form a composite sample. Three 50 cm × 50 cm quadrats were established to collect aboveground biomass (i.e., the sum of leaves, stems and litter), and three root sampling blocks (15-cm length × 15-cm width × 30 cm depth) were excavated to collect root biomass in each community of each transect. All soil and plant samples were stored at 4 °C in the field and then transported to the laboratory for subsequent analysis.
Laboratory analysis
Each root sampling block was put through a 100 mesh sieve and flushed with water, and the roots remaining in the sieve were collected at the final step14. All plant samples were carefully cleaned and oven-dried at 65 °C for determining aboveground and root biomass. The visible plant and fauna residues were removed from the soil samples, and soil samples were then divided into three subsamples after thorough mixing. One subsample was air-dried and passed through 1-mm sieves to measure soil pH, salinity, SOC and SON. A subsample of 2-mm sieved fresh soil was stored at 4 °C to determine WSOC, MBC, MBN and microbial respiration. Another subsample was passed through 2-mm sieves and stored at −80 °C as quickly as possible after freeze-drying and was used to analyze for PLFAs. The soil subsample was weighed and oven-dried at 105 °C to determine soil moisture. Soil pH was measured in a soil–water suspension (1:2.5 soil:water) with a glass electrode. Soil salinity was determined in a soil–water suspension (1:5 soil:water) with a conductivity meter. Before the SOC and SON analyses, approximately 10 g of dried soil subsamples were treated with 1 M HCl at room temperature for 24 h to eliminate total inorganic C and N29, and unhydrolyzed residues were analyzed with a CN elemental analyzer (Vario PYRO cube elemental analyzer, Germany) to obtain SOC and SON concentrations14. WSOC was determined using the method described by Yang et al.16. Briefly, WSOC was extracted from 10 g moist soil samples after addition of 20 mL distilled water. The extracted fluid was vacuum filtered through a 0.45 μm filter, and C concentration of the filtrate was rapidly determined by a Liqui TOCII analyzer (Elementar Analysensystem GmbH, Germany).
MBC and MBN were measured using the chloroform fumigation-extraction method51. 25 g dry-weight-equivalent of moist soil was fumigated with ethanol-free chloroform for 48 h at 25 °C in the dark. The fumigated and un-fumigated samples were then extracted with 100 mL 0.5 M K2SO4 by shaking for 30 min at 200 rpm and then filtered. Organic C and TN in the K2SO4-extracted solution were determined using a Liqui TOCII analyzer and the Kjeldahl method, respectively. MBC and MBN were calculated according to the equation: MBC = Ec/0.38, MBN = En/0.54, where Ec and En were organic C and TN extracted from fumigated soil subtracted organic C and TN extracted from unfumigated soil, respectively.
Phospholipid fatty acids analysis
The soil microbial community composition was assessed using PLFAs analysis based on the method of Bossio and Scow52. Briefly, lipids were extracted from 8 g of a dry-weight-equivalent of the fresh soil subsample using 23 mL of an extraction mixture of chloroform: methanol: phosphate buffer (1:2:0.8, v/v/v). Phospholipids were then split into neutral, glyco- and phospho- lipids using solid-phase extraction columns by eluting with CHCl3, acetone and methanol, respectively. Subsequently, phospholipids were subjected to a mild-alkali methanolysis to recover fatty acid methyl esters. Samples were then re-dissolved in 200 ml hexane containing nonadecanoic acid methyl ester (19:0) as an internal standard and were analyzed using a Hewlett-Packard 6890 Gas Chromatograph equipped with an Ultra 2-methylpolysiloxane column with N2 as the carrier gas and H2 and air to support the flame. A 2-μL injection of the above dilution with a 1:50 split was employed at 250 °C for the injector and 300 °C for the detector. The oven temperature ramped from 170 °C to 300 °C at 5 °C min−1 and was held for 12 min. Peaks were identified using bacterial fatty acid standards and MIDI peak identification software (MIDI, Newark, DE). The quantity (ng g−1 dry soil) of each PLFAs was calculated based on the 19:0 internal standard (5 μg mL−1). The quantities of the PLFAs in each sample were expressed as ng PLFAs g−1 dry soil and were used to estimate microbial biomass. Bacteria were represented by the sum of the PLFAs: i14:0, i15:0, a15:0, 15:0, i16:0, i17:0, a17:0, 17:0, cy17:0, 14:1ω5c, 15:1ω6c, 16:1ω7c, and 18:1ω7c44,52,53. Fungi were represented by the sum of the PLFAs: 18:1ω9c, 18:2ω6,9c, and 20:1ω9c53,54,55. Gram+ bacteria were identified by the PLFAs: i13:0, i14:0, i15:0, a15:0, i16:0, a16:0, i17:0, and a17:0, and gram− bacteria were identified by the PLFAs: 14:1ω5c, 15:1ω6c, 16:1ω7, 16:1ω9c, 17:1ω8c, 18:1ω7c, 12:0 2OH, 15:0 3OH, 16:1 2OH, cy17:0, cy19:0 ω8c, and 18:1ω7c 11-methyl44,54,56. AMF were identified by the PLFAs 16:1ω5c44,54,57. Actinomycete were identified by the PLFAs 10me 16:0 and 10me 17:041. Monounsaturated PLFAs were estimated from the sum of the following PLFAs: 14:1ω5c, 15:1ω6c, 16:1ω5c, 16:1ω7c, 16:1ω9c, 17:1ω8c, 18:1ω7c, 18:1ω9c, and 20:1ω9c21,41,44. Branched PLFAs were estimated from the sum of following PLFAs: i13:0, i14:0, i15:0, a15:0, i16:0, a16:0, i17:0, a17:0, 10me 16:0, 10me 17:0, 12:0 2OH, 15:0 3OH, and16:1 2OH41,44,52. SSC PLFAs were estimated from the sum of the PLFAs: 12:0, 13:0, 14:0, 15:0, 16:0, 17:0, 18:0, and 20:044,52. The total PLFAs of the microbial community were represented by the sum of fungal PLFAs, gram+ bacterial PLFAs, gram− bacterial PLFAs, AMF PLFAs, actinomycete PLFAs, SSC PLFAs, and 20:4ω6,9,12,15c. The ratios of fungal:bacterial PLFAs, gram−:gram+ and monounsaturated:branched PLFAs were calculated from the above PLFAs. The ratio of cy17:0/16:1ω7c was used as a bacterial stress index, which indicates the growth stage of the gram− bacteria community58.
Microbial respiration measurements
Microbial respiration was measured by alkali absorption of CO2 evolved at 25 °C and 35 °C for 30 days in a laboratory aerobic incubation experiment with soil37,48. Briefly, the fresh soil sample (20 g dry weight equivalent) was evenly placed in a 50 mL glass beaker. Distilled water was added to the soil samples to maintain moisture at 60% of water-holding capacity. The glass beaker was placed in a 500 mL mason jar, and the glass tubes containing 10 mL 0.5 M NaOH solution was placed in each mason jar to capture CO2 evolved by the soil in the mason jar. The mason jar was sealed and incubated at 25 °C and 35 °C in the dark for 30 days. After incubation for 6, 12, 18, 24, and 30 days, the glass tubes that were equipped with NaOH were removed, and the mason jar was opened for several minutes to maintain sufficient O2 levels. The amount of CO2 was determined by titration of the NaOH solution with 0.1 M HCl in two drops BaCl2.
Temperature sensitivity (Q10) of microbial respiration was determined using equation (1)48,59.
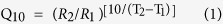 |
where R2 and R1 are the mean microbial respiration rate at T2 (35 °C) and T1 (25 °C), which are the temperature levels within 30 days of incubation.
The microbial respiration quotient (qCO2) was calculated by dividing the microbial respiration (mg CO2 30 day) per kg by the MBC36.
Statistical analyses
All of the statistical analyses were performed using SPSS Statistics 19 software. Data not meeting assumptions of normality and homogeneity of variance were log- or cube root-converted prior to statistical testing. One-way analysis of variance (ANOVA) was used to determine the statistical significance of the effect of S. alterniflora invasion time on soil and plant properties, microbial biomass and various types of PLFAs, cumulative microbial respiration, microbial respiration:total PLFAs ratio, qCO2 and Q10. One-way ANOVA was also used to determine the statistical significance of the incubation temperatures on cumulative microbial respiration, the ratio of microbial respiration:total PLFAs and qCO2. Pearson’s correlation analysis was performed to correlate soil microbial indexes with the soil and plant characteristics, and to correlate soil microbial respiration indexes with microbial biomass (i.e., each of the PLFAs). Soil and plant characteristics were tested for significant contributions to explain the variations in the PLFAs data with redundancy analysis (RDA) using CANOCO software for Windows 4.5. The statistical significance of the RDA was tested using the Monte Carlo permutation test (499 permutations; P < 0.05).
Additional Information
How to cite this article: Yang, W. et al. Spartina alterniflora invasion alters soil microbial community composition and microbial respiration following invasion chronosequence in a coastal wetland of China. Sci. Rep. 6, 26880; doi: 10.1038/srep26880 (2016).
Acknowledgments
This work was supported by the National Basic Research Program of China (grant no. 2013CB430400) and the Strategic Priority Research Program B of the Chinese Academy of Sciences (XDB15010200). We are grateful to Zhihui Shi for assistance with the fieldwork, Prof. Lixia Zhou for analysis and identification of PLFAs, and all of the members of the Jiangsu Yancheng Wetland National Nature Reserve for Rare Birds for supporting this work. Last but not least, we appreciate anonymous reviewers and editor for their valuable comments and precious suggestions on this paper.
Footnotes
Author Contributions W.Y., S.Q.A. and X.L.C. designed the research. W.Y. conducted the experiment, analysed the data, and drafted the manuscript. N.J. helped carry out the laboratory analyses. X.L. helped interpret the results of the study. W.Y. and X.L.C. contributed substantially to revisions.
References
- Pimentel D., Lach L., Zuniga R. & Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience 50, 53–65 (2000). [Google Scholar]
- Didham R. K., Tylianakis J. M., Hutchison M. A., Ewers R. M. & Gemmell N. J. Are invasive species the drivers of ecological change? Trends Ecol. Evol. 20, 470–474 (2005). [DOI] [PubMed] [Google Scholar]
- Mack R. N. et al. Biotic invasions: Causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710 (2000). [Google Scholar]
- Liao C. Z. et al. Altered ecosystem carbon and nitrogen cycles by plant invasion: a meta-analysis. New phytol. 177, 706–714 (2008). [DOI] [PubMed] [Google Scholar]
- van der Putten W. H. et al. Plant–soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276 (2013). [Google Scholar]
- van der Putten W. H., Klironomos J. N. & Wardle D. A. Microbial ecology of biological invasions. ISME J. 1, 28–37 (2007). [DOI] [PubMed] [Google Scholar]
- Inderjit & van der Putten W. H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 25, 512–519 (2010). [DOI] [PubMed] [Google Scholar]
- Elgersma K. J., Ehrenfeld J. G., Yu S. & Vor T. Legacy effects overwhelm the short-term effects of exotic plant invasion and restoration on soil microbial community structure, enzyme activities, and nitrogen cycling. Oecologia 167, 733–745 (2011). [DOI] [PubMed] [Google Scholar]
- Hawkes C. V., Wren I. F., Herman D. J. & Firestone M. K. Plant invasion alters nitrogen cycling by modifying the soil nitrifying community. Ecol. Lett. 8, 976–985 (2005). [DOI] [PubMed] [Google Scholar]
- Rodgers V. L., Wolfe B. E., Werden L. K. & Finzi A. C. The invasive species Alliaria petiolata (garlic mustard) increases soil nutrient availability in northern hardwood-conifer forests. Oecologia 157, 459–471 (2008). [DOI] [PubMed] [Google Scholar]
- Kao-Kniffin J. & Balser T. C. Soil fertility and the impact of exotic invasion on microbial communities in Hawaiian forests. Microb. Ecol. 56, 55–63 (2008). [DOI] [PubMed] [Google Scholar]
- Liao J. D. & Boutton T. W. Soil microbial biomass response to woody plant invasion of grassland. Soil Biol. Biochem. 40, 1207–1216 (2008). [Google Scholar]
- Wilsey B. J. & Polley H. W. Aboveground productivity and root-shoot allocation differ between native and introduced grass species. Oecologia 150, 300–309 (2006). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Labile and recalcitrant soil carbon and nitrogen pools in tidal salt marshes of the eastern Chinese coast as affected by short-term C4 plant Spartina alterniflora invasion. Clean–Soil, Air, Water 43, 872–880 (2015). [Google Scholar]
- Lazzaro L. et al. Soil and plant changing after invasion: The case of Acacia dealbata in a Mediterranean ecosystem. Sci. Total Environ. 497–498, 491–498 (2014). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Consequences of short-term C4 plant Spartina alterniflora invasions for soil organic carbon dynamics in a coastal wetland of Eastern China. Ecol. Eng. 61, 50–57 (2013). [Google Scholar]
- Wan X. L. et al. Soil C:N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations. Plant Soil 387, 103–116 (2015). [Google Scholar]
- Högberg M. N., Högberg. P. & Myrold. D. D. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150, 590–601 (2007). [DOI] [PubMed] [Google Scholar]
- Yuan J. J., Ding W. X., Liu D. Y., Xiang J. & Lin Y. X. Methane production potential and methanogenic archaea community dynamics along the Spartina alterniflora invasion chronosequence in a coastal salt marsh. Appl. Microbiol. Biot. 98, 1817–1829 (2014). [DOI] [PubMed] [Google Scholar]
- Yuan J. J. et al. Exotic Spartina alterniflora invasion alters ecosystem–atmosphere exchange of CH4 and N2O and carbon sequestration in a coastal salt marsh in China. Global Change Biol. 21, 1567–1580 (2015). [DOI] [PubMed] [Google Scholar]
- Kourtev P. S., Ehrenfeld J. G. & Häggblom M. Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 35, 895–905 (2003). [Google Scholar]
- Chen D. M. et al. Tree girdling affects the soil microbial community by modifying resource availability in two subtropical plantations. Appl. Soil Ecol. 53, 108–115 (2012). [Google Scholar]
- Banerjee S. et al. Legacy effects of soil moisture on microbial community structure and N2O emissions. Soil Biol. Biochem. 95, 40–50 (2016). [Google Scholar]
- Fierer N., Schimel J. P. & Holden P. A. Influence of drying-rewetting frequency on soil bacterial community structure. Microb. Ecol. 45, 63–71 (2003). [DOI] [PubMed] [Google Scholar]
- Fierer N. & Jackson R. B. The diversity and biogeography of soil bacterial communities. P. Natl. Acad. Sci. USA 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. P. et al. Changes in the soil microbial community structure with latitude in eastern China, based on phospholipid fatty acid analysis. Appl. Soil Ecol. 43, 234–240 (2009). [Google Scholar]
- Rousk J., Brookes P. C. & Bååth E. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol. Biochem. 42, 516–520 (2010). [Google Scholar]
- Kamble P. N., Gaikwad V. B., Kuchekar S. R. & Bååth E. Microbial growth, biomass, community structure and nutrient limitation in high pH and salinity soils from Pravaranagar (India). Eur. J Soil Biol. 65, 87–95 (2014). [Google Scholar]
- Cheng X. L. et al. Short-term C4 plant Spartina alterniflora invasions change the soil carbon in C3 plant-dominated tidal wetlands on a growing estuarine Island. Soil Biol. Biochem. 38, 3380–3386 (2006). [Google Scholar]
- Huang J. X. et al. Responses of soil nitrogen fixation to Spartina alterniflora invasion and nitrogen addition in a Chinese salt marsh. Sci. Rep. 6, 20384, doi: 10.1038/srep20384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C. Z. et al. Invasion of Spartina alterniflora enhanced ecosystem carbon and nitrogen stocks in the Yangtze Estuary, China. Ecosystems 10, 1351–1361 (2007). [Google Scholar]
- Vries F. T. et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 15, 1230–1239 (2012). [DOI] [PubMed] [Google Scholar]
- Haynes R. J. Labile organic matter as an indicator of organic matter quality in arable and pastoral soils in New Zealand. Soil Biol. Biochem. 32, 211–219 (2000). [Google Scholar]
- Vanhala P. et al. Temperature sensitivity of soil organic matter decomposition in southern and northern areas of the boreal forest zone. Soil Biol. Biochem. 40, 1758–1764 (2008). [Google Scholar]
- Bailey V. L., Smith J. L. & Bolton H. Jr. Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol. Biochem. 34, 997–1007 (2002). [Google Scholar]
- Huang Y. M., Liu D. & An S. S. Effects of slope aspect on soil nitrogen and microbial properties in the Chinese Loess region. Catena 125, 135–145 (2015). [Google Scholar]
- Zhang W. et al. Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Global Change Biol. 11, 266–277 (2005). [Google Scholar]
- Joergensen R. G. & Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 40, 2977–2991 (2008). [Google Scholar]
- van der Heijden M. G. A., Bardgett R. D. & van Straalen N. M. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008). [DOI] [PubMed] [Google Scholar]
- Cusack D. F., Silver W. L., Torn M. S., Burton S. D. & Firestone M. K. Change in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 92, 621–632 (2011). [DOI] [PubMed] [Google Scholar]
- Bossio D. A., Fleck J. A., Scow K. M. & Fujii R. Alteration of soil microbial communities and water quality in restored wetlands. Soil Biol. Biochem. 38, 1223–1233 (2006). [Google Scholar]
- Smith A. P., Marín-Spiotta E., de Graaff M. A. & Balser T. C. Microbial community structure varies across soil organic matter aggregate pools during tropical land cover change. Soil Biol. Biochem. 77, 292–303 (2014). [Google Scholar]
- Li N. et al. Fungi contribute more than bacteria to soil organic matter through necromass accumulation under different agricultural practices during the early pedogenesis of a Mollisol. Eur. J. Soil Biol. 67, 51–58 (2015). [Google Scholar]
- Cao Y. S. et al. Soil microbial community composition under Eucalyptus plantations of different age in subtropical China. Eur. J. Soil Biol. 46, 128–135 (2010). [Google Scholar]
- Gorham E. Northern peatlands: role in the carbon cycle and probable responses to climate warming. Ecol. Appl. 1, 182–195 (1991). [DOI] [PubMed] [Google Scholar]
- Whitting G. J. & Chanton J. P. Greenhouse carbon balance of wetlands: methane emission versus carbon sequestration. Tellus B 53, 521–528 (2001). [Google Scholar]
- Yuste J. C. et al. Microbial soil respiration and its dependency on carbon inputs, soil temperature and moisture. Global Change Biol. 13, 2018–2035 (2007). [Google Scholar]
- Nie M. et al. Positive climate feedbacks of soil microbial communities in a semi-arid grassland. Ecol. Lett. 16, 234–241 (2013). [DOI] [PubMed] [Google Scholar]
- Wardle D. A. & Ghani A. A critique of the microbial metabolic quotient (qCO2) as a bio-indicator of disturbance and ecosystem development. Soil Biol. Biochem. 27, 1601–1610 (1995). [Google Scholar]
- Belay-Tedla A., Zhou X., Su B., Wan S. & Luo Y. Labile, recalcitrant, and microbial carbon and nitrogen pools of a tallgrass prairie soil in the US Great Plains subjected to experimental warming and clipping. Soil Biol. Biochem. 41, 110–116 (2009). [Google Scholar]
- Vance E. D., Brookes P. C. & Jenkinson D. S. An extraction method for measuring microbial biomass C. Soil Biol. Biochem. 19, 703–707 (1987). [Google Scholar]
- Bossio D. A. & Scow K. M. Impacts of carbon and flooding on soil microbial communities: phospholipid fatty acid profiles and substrate utilization patterns. Microb. Ecol. 35, 265–278 (1998). [DOI] [PubMed] [Google Scholar]
- Bååth E. & Anderson T. H. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35, 955–963 (2003). [Google Scholar]
- Kourtev P. S., Ehrenfeld J. G. & Häggblom M. Exotic plant species alter the microbial community structure and function in the soil. Ecology 83, 3152–3166 (2002). [Google Scholar]
- Swallow M., Quideau S. A., MacKenzie M. D. & Kishchuk B. E. Microbial community structure and function: the effect of silvicultural burning and topographic variability in northern Alberta. Soil Biol. Biochem. 41, 770–777 (2009). [Google Scholar]
- Sampedro L., Jeannotte R. & Whalen J. K. Trophic transfer of fatty acids from gut microbiota to the earthworm Lumbricus terrestris L. Soil Biol. Biochem. 38, 2188–2198 (2006). [Google Scholar]
- Kong C. H., Wang P., Zhao H., Xu X. H. & Zhu Y. D. Impact of allelochemical exuded from allelopathic rice on soil microbial community. Soil Biol. Biochem. 40, 1862–1869 (2008). [Google Scholar]
- Ponder F. & Tadros M. Phospholipid fatty acids in forest soil four years after organic matter removal and soil compaction. Appl. Soil Ecol. 19, 173–182 (2002). [Google Scholar]
- Davidson E. A. & Janssens I. A. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173 (2006). [DOI] [PubMed] [Google Scholar]