Abstract
Background:
This meta-analysis was conducted to evaluate whether HTR1A gene polymorphisms impact the efficacy of antipsychotic drugs in patients with schizophrenia.
Methods:
Candidate gene studies that were published in English up to August 6, 2015 were identified by a literature search of PubMed, Web of Science, and Google scholar. Data were pooled from individual clinical trials considering overall symptoms, positive symptoms and negative symptoms, and standard mean differences were calculated by applying a random-effects model.
Results:
The present meta-analysis included a total of 1281 patients from 10 studies. Three polymorphisms of HTR1A (rs6295, rs878567, and rs1423691) were selected for the analysis. In the pooled data from all studies, none of these HTR1A polymorphisms correlated significantly with either overall symptoms or positive symptoms. However, C allele carriers of the rs6295 polymorphism showed a significantly greater negative symptoms improvement than G allele carriers (P=.04, standardized mean difference =-0.14, 95%CI = 0.01 to 0.28).
Conclusions:
The results of our present analysis indicate that the HTR1A rs6295 polymorphism may impact negative symptoms improvement but not on either overall symptoms or positive symptoms improvement. However, this meta-analysis was based on a small number of studies and patients, and the effect size on negative symptoms was small. Given this limitation, the results should be confirmed by further investigations.
Keywords: HTR1A, polymorphism, schizophrenia, antipsychotics, efficacy
Introduction
The effects of antipsychotic medications in schizophrenia are mediated by their interactions with several receptors expressed in the brain. Currently, the dopamine D2 and the serotonin 2A receptor are assumed to be representative target receptors of antipsychotic drugs. The serotonin 1A receptor (5-HT1AR) is another important receptor, since it has been implicated in both schizophrenia pathogenesis and antipsychotic mechanisms of action. 5-HT1AR is expressed in various sites throughout the brain such as the hippocampus (Barnes and Sharp, 1999; Aznar et al., 2003; Varnas et al., 2004), amygdala, and hypothalamus. A postmortem study including patients with schizophrenia demonstrated elevated 5-HT1AR density in the cortex of the frontal lobe (Hashimoto et al., 1991; Burnet et al., 1996; Sumiyoshi et al., 1996; Tauscher et al., 2002). This might be explained as an effect of serotonergic hypo-activation and glutamatergic hypo-transmission in this region (Wedzony et al., 1997; Newman-Tancredi and Kleven, 2011). In addition, second-generation antipsychotics, including aripiprazole, bifeprunox, clozapine, lurasidone, perospirone, quetiapine, and ziprasidone, are hypothesized to exhibit partial agonistic actions on the 5-HT1AR, and molecules directly targeting the 5-HT1AR ameliorate psychiatric symptoms (Jordan et al., 2002; Odagaki and Toyoshima, 2007; Murasaki et al., 2008; Fagiolini et al., 2010; Lerond et al., 2013). Furthermore, adjunctive therapy with anxiolytic agents exerting a partial agonistic action on the 5-HT1AR, such as buspirone and tandospirone, is known to improve psychiatric symptoms in schizophrenic patients (Kishi et al., 2013a). Based on these observations, the action of antipsychotics on the 5-HT1AR is supposed to play an important role in the treatment of psychiatric disorders.
The HTR1A gene encoding 5-HT1AR, mapped on chromosome 5q11.2–13, is an intronless gene approximately 2200bp in size, including an approximately 1200-bp coding sequence known to code for 422 amino acids (http://www.ncbi.nlm.nih.gov/nuccore). A number of single nucleotide polymorphisms (SNPs) have been identified for HTR1A; a particularly important SNP is rs6295 (C-1019G), which is located in the promoter region and is known to be a functional polymorphism that regulates HTR1A transcription and region-specific modification of HTR1A expression (Wu and Comings, 1999; Lemonde et al., 2003; Albert and Lemonde, 2004). Thus, pharmacogenetic candidate gene studies were mainly focused on rs6295, and they investigated the relationship between psychiatric drug efficacy in mental disorders and rs6295, with the aim of developing personalized treatments. Several meta-analyses investigated data from studies focused on antidepressant efficacy and the rs6295 polymorphism, but none documented a statistically significant relationship on all populations or on Caucasian/Asian populations (Kato and Serretti, 2010; Zhao et al., 2012; Niitsu et al., 2013). Previous studies identified an association between the rs6295 polymorphism and symptomatic improvement in schizophrenia during treatment with antipsychotics, taking into consideration overall improvement of symptoms (Reynolds et al., 2006), amelioration of negative symptoms (Reynolds et al., 2006; Wang et al., 2008; Mossner et al., 2009), and improved attention (Sumiyoshi et al., 2010). On the other hand, negative findings exist (Ikeda et al., 2008; Crisafulli et al., 2012; Tang et al., 2014; Takekita et al., 2015); thus, no clear conclusion can be traced about the effect of this SNP on antipsychotic efficacy. Other HTR1A gene polymorphisms (in particular rs878567 and rs1423691) were investigated in regard to antipsychotic response, even if no functional effect is known for them. Some studies suggested that they may be associated with antipsychotic efficacy (Crisafulli et al., 2012; Gupta et al., 2012; Drago et al., 2013; Takekita et al., 2015).
However, the results obtained to date have not been comprehensively assessed in a meta-analysis. Thus, the present meta-analysis aimed to analyze the cumulative knowledge provided by candidate gene studies focused on the association between HTR1A gene polymorphisms and antipsychotic efficacy in schizophrenia.
Methods
This meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (Moher et al., 2009).
Search
We conducted a literature search using PubMed, Web of Science, and Google scholar to identify articles published in English until August 6, 2015. Search queries used were combinations of the following phrases: “schizophrenia or psychosis,” “serotonin 1A receptor or 5-HT 1A, HTR1A,” “antipsychotics or antipsychotic agents,” “response or efficacy,” and “gene or polymorphism.” In this search, the generic name of each antipsychotic was also used in place of “antipsychotics or antipsychotic agents.”
Furthermore, we also manually searched related journals published in English to complement the electronic search. Two independent investigators (Y.T., Y.K.) conducted the literature search.
Inclusion Criteria, Data Extraction, and Outcomes
The following inclusion criteria were applied to select includible studies: (1) They investigated the association between HTR1A polymorphisms and antipsychotic clinical efficacy; (2) The majority of patients had a diagnosis of schizophrenia or schizoaffective disorder based on DSM or ICD criteria (not applicable to patients with first psychotic episode); (3) Drug response was assessed using a standardized rating scale, such as the Positive and Negative Syndrome Scale or the Clinical Global Impression, at baseline and follow-up evaluations; (4) Genotype distribution was in Hardy–Weinberg equilibrium; (5) The study was conducted using the candidate gene approach; (6) The study was published in English and in a peer-reviewed journal; and (7) The study was based on an independent sample (nonoverlapping with other studies). When outcome data were not available in the article, we requested data from the author or we extracted the data from the published figures by the GSYS 2.4 program (Japan Charged-Particle Nuclear Reaction Data Group, http://www.jcprg.org/gsys/2.4/index-j.html, Sapporo, Japan). Based on the data obtained from articles that met the above criteria, we selected HTR1A SNPs with at least 3 includible studies. Two authors (Y.T. and Y.K.) independently extracted and checked the data. Any disagreement was resolved by discussion until consensus was reached.
The purposes of the present meta-analysis were to identify possible associations between HTR1A SNPs and baseline to endpoint changes in: overall symptoms, positive symptoms, negative symptoms.
Data Analysis
Data were analyzed using a random-effects model as defined by DerSimonian and Laird (1986), because clinical heterogeneity among the populations and interventions included was expected. The standardized mean difference (SMD) and its 95% CI were calculated for continuous outcomes. Missing SDs were imputed according to the method described in Section 16.1.3 of the Cochrane Handbook for Systematic Reviews of Interventions (The Cochrane Collaboration, 2011). Study heterogeneity was measured using the chi-squared and I-squared statistics, with chi-squared values of P < .05 and I2 values of >50% indicating relevant heterogeneity (Higgins et al., 2003). In addition to the main analysis, we performed a subgroup analysis defined a priory by race, that is, Asians vs Caucasians.
Furthermore, we performed a meta-regression using age (mean age for each sample), sex (percentage of males in each study), and study duration as the moderator variables. Publication bias was assessed by constructing funnel plots and conducting an Egger’s test (Egger et al., 1997).
Data were analyzed using Review Manager 5.3.5 (Cochrane Informatics & Knowledge Management Department, http://tech.cochrane.org/revman). Stata/IC 14.0 (Stata Corporation, College Station, TX, USA) was used to perform the Egger’s test and meta-regression analyses.
Results
Included Studies
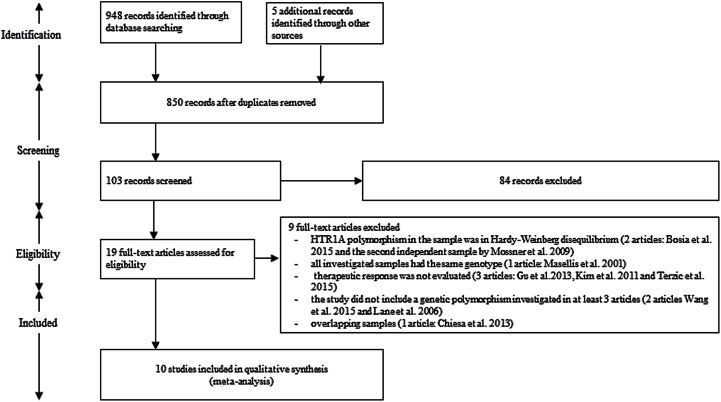
The electronic search yielded a total of 948 potential studies. In addition, we found 5 studies when conducting the manual search. Among the 953 identified hits, 850 articles were found to be duplicated between the databases. Of the 103 unique studies, we excluded 84 articles based on title and abstract reviews. Based on full-text inspection, we excluded 9 other references for the following reasons: the HTR1A polymorphism investigated in the sample was not in Hardy-Weinberg equilibrium (2 articles: Bosia et al., 2015 and Mossner et al., 2009); all the investigated samples had the same genotype (1 article: Masellis et al., 2001); therapeutic response was not evaluated (3 articles: Kim and Yoon, 2011; Gu et al., 2013; Terzic et al., 2015); the study did not include a polymorphism investigated by at least 2 other articles (2 articles: Lane et al., 2006; Wang et al., 2015); overlapping samples (1 article: Chiesa et al., 2013).
Ultimately, 10 studies including 1281 patients were included in the present meta-analysis (Reynolds et al., 2006; Ikeda et al., 2008; Wang et al., 2008; Mossner et al., 2009; Sumiyoshi et al., 2010; Crisafulli et al., 2012; Gupta et al., 2012; Drago et al., 2013; Tang et al., 2014; Takekita et al., 2015). Three HTR1A polymorphisms were included in the analysis (rs6295 investigated by 10 studies, rs878567 by 4 studies, and rs1423691 by 3 studies). Figure 1 shows the flowchart of study selection and inclusion. Table 1 summarizes the characteristics of the included studies. Each study included 30 to 398 patients (median: 98) with a mean age of 32.4 years and a study duration of 4 weeks to 3 months. Male patients constituted 53.9% of the sample population, with Asians, Caucasians, and South Indians accounting for 53.3%, 17.7%, and 29.0%, respectively. Eight studies included only patients who had been diagnosed with schizophrenia; 3 of these studies were conducted in patients experiencing first-episode schizophrenia. Three studies used a single antipsychotic agent; in 2 of these studies that agent was risperidone. Eight studies used the Positive and Negative Syndrome Scale score as a parameter for assessing psychiatric symptoms, while the others used the Clinical Global Impression-I, SAPS, or SANS score. In all studies, genotype distributions of rs6295, rs878567, and rs1423691 were in Hardy–Weinberg equilibrium. No subgroup analysis or meta-regression was performed for polymorphisms other than rs6295, due to the limited number of studies evaluating these polymorphisms.
Figure 1.
Study flow diagram.
Table 1.
Description of the Included Studies
| Study | Sample Size (male/female) | Country | Ethnicity |
Age
(SD) |
Setting | Diagnostic Criteria | Patient Type | Medication | Duration of Treatment | Outcome Measure | HTR1A SNP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crisafulli et al., 2012 | 221 (126/95) |
Korea | Asian | 38.01 (12.67) |
In-patients | DSM-IV | Not any particular restriction | RIS, OLZ, QEP, AMI and others (All APDs are not identified) | 37.64±1S6.76 days from admission to discharge | PANSS | rs10042486, rs6295, rs878567, rs1423691 |
| Drago et al., 2013 | 96 (51/45) |
German, Yugoslavia, Turkey, Other | Caucasian | 34.28 (11.29) |
In-patients | DSM-IV | Acute | HPD | 4W | PANSS | rs6295, rs878567, rs1423691, rs10085024 |
| Gupta et al., 2012 | 371 (224/147) |
India | South Indian | 29.63 (8.63) |
In and out- patients | DSM-IV | Most of them are of chronic patients | RIS, OLZ, CLZ, ZIP, QEP, ARI, AMI | 3M | CGI-I | rs6295, rs34118353, rs878567, rs1423691 |
| Ikeda et al., 2008 | 120 (58/62) |
Japan | Asian | 31.2 (8.7) |
In and out- patients | DSM-IV-TR | First-episode, neuroleptic- naïve DUP < 5 y |
RIS | 8W | PANSS | rs6295 |
| Mossner et al., 2009 | 68 (44/24) |
German | Caucasian | 31.7 | NR | ICD-10 | First-episode | RIS, HPD | 4W | PANSS | rs6295 |
| Reynolds et al., 2006 | 63 (45/18) |
Spain | Caucasian | 25.1 (6.5) |
In and out- patients | Drug-naive patients with first-episode psychosis | Drug-naive patients with first-episode psychosis | RIS, OLZ, QEP, HPD, ZIP, AMI | 3M | PANSS | rs6295 |
| Sumiyoshi et al., 2010 | 30 (11/19) |
Japan | Asian | 31.6 (11.0) |
Out-patients | DSM-IV-TR | Acute | OLZ, PER | 3M | SAPS, SANS | rs6295 |
| Takekita et al., 2015 | 100 (43/57) |
Japan | Asian | 44.1 (16.2) |
In and out- patients | DSM-IV-TR | Acute | PER, ARI | 12W | PANSS | rs1364043, rs878567, rs6295, rs10042486 |
| Tang et al., 2014 | 82 (45/37) |
Chinese | Asian | 25.8 (7.1) |
In-patients | DSM-IV | First psychotic episode | Baseline:CPZ, RIS, CLZ, FLU endpoint:CLZ, CPZ, RIS |
10W | PANSS and PANSS 5 factors | rs6295 |
| Wang et al., 2008 | 130 (43/87) |
China | Asian | 30.35 (11.33) |
NR | DSM-IV | - Not treatment resistance - SGAs-naïve - Not APDs administration for 4W |
RIS | 8W | PANSS | rs6295 |
Abbreviations: AMI, amisulpride; APD, antipsychotic drug; ARI, aripiprazole; CGI-I, clinical global impression of improvement; CLZ, clozapine; CPZ, chlorpromazine; DUP, duration of untreated psychosis; FLU, fluphenazine; HPD, haloperidol; NR, not reported; OLZ, olanzapine; PANSS, Positive and Negative Syndrome Scale; PER, perospirone; QEP, quetiapine; RCT, randomized clinical trial; RIS, risperidone; SAPS, scale for assessment of positive symptoms; SANS, scale for the assessment of negative symptoms; SGA, second-generation antipsychotic; SNP, single nucleotide polymorphism; ZIP, ziprasidone.
rs6295 and Antipsychotic Drug Response
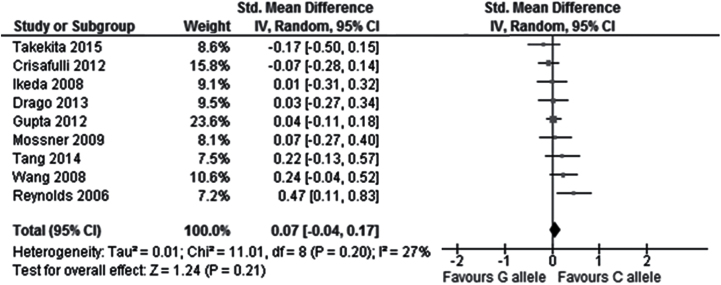
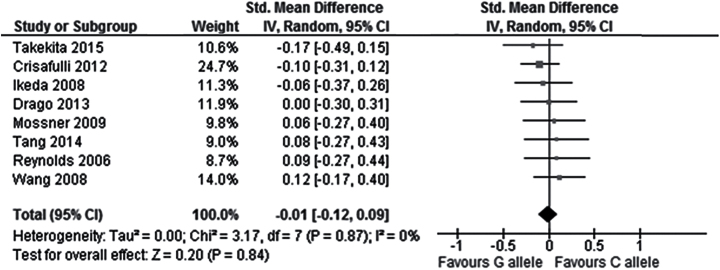
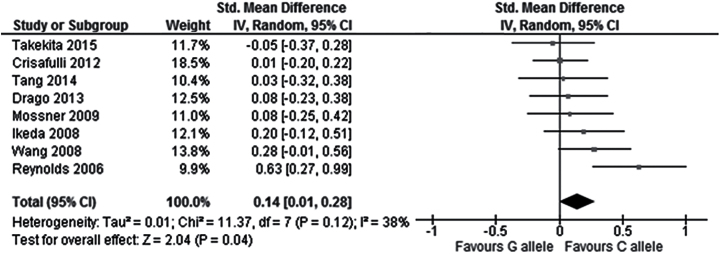
In the total population, no significant correlation was observed between the rs6295 polymorphism and overall symptoms change from baseline to endpoint in the comparison between CC and G carriers (9 studies, SMD = 0.08, Cl = -0.09 to 0.25, P=.37), the comparison between C and GG carriers (9 studies, SMD = 0.11, CI = -0.06 to 0.27, P=.20), or the comparison between the C allele and the G allele (9 studies, SMD = 0.07, CI = -0.04 to 0.17, P=.21; Figure 2; Table 2). Similarly, no significant correlation was found between the rs6295 polymorphism and positive symptoms change in the comparison CC vs G carriers (9 studies, SMD = 0.02, Cl = -0.12 to 0.16, P=.76), C vs GG carriers (8 studies, SMD = -0.04, CI = -0.26 to 0.18, P=.73), or C vs G allele (8 studies, SMD = -0.01, CI = -0.12 to 0.09, P=.84; Figure 3; Table 2). Similarly, no significant correlation was found between the rs6295 polymorphism and negative symptoms change in the comparison CC vs G carriers (9 studies, SMD = 0.18, Cl = -0.04 to 0.39, P=.10) or the comparison C vs GG carriers (8 studies, SMD = 0.13, CI = -0.09 to 0.36, P=.24), but a significantly higher improvement in negative symptoms was observed in C allele carriers when comparing C vs G allele (8 studies, SMD =-0.14, CI = 0.01 to 0.28, P=.04) (Figure 4; Table 2).
Figure 2.
Forest plot for the association with rs6295 polymorphism (C alleles vs G alleles) and improvement of overall symptoms.
Table 2.
Association between the HTR1A polymorphisms and antipsychotic efficacy
| N | N | SMD | 95 % CI | Test for Overall Effect | Heterogeneity | P Value of Egger’s Test | |||
|---|---|---|---|---|---|---|---|---|---|
| P | Z | P | I2 (%) | ||||||
| rs6295 | |||||||||
| Overall symptoms | |||||||||
| CC vs G carriers | 9 | 1226 | 0.08 | -0.09 to 0.25 | .37 | 0.90 | .06 | 47 | .243 |
| C carriers vs GG | 9 | 1226 | 0.11 | -0.06 to 0.27 | .20 | 1.28 | .78 | 0 | .823 |
| C alleles vs G alleles | 9 | 2452 | 0.07 | -0.04 to 0.17 | .21 | 1.24 | .20 | 27 | .352 |
| Positive symptoms | |||||||||
| CC vs G carriers | 9 | 894 | 0.02 | -0.12 to 0.16 | .76 | 0.30 | .67 | 0 | .112 |
| C carriers vs GG | 8 | 864 | -0.04 | -0.26 to 0.18 | .73 | 0.35 | .96 | 0 | .309 |
| C alleles vs G alleles | 8 | 1728 | -0.01 | -0.12 to 0.09 | .84 | 0.20 | .87 | 0 | .26 |
| Negative symptoms | |||||||||
| CC vs G carriers | 9 | 894 | 0.18 | -0.04 to 0.39 | .10 | 1.62 | .03* | 53 | .292 |
| C carriers vs GG | 8 | 864 | 0.13 | -0.09 to 0.36 | .24 | 1.17 | .55 | 0 | .727 |
| C alleles vs G alleles | 8 | 1728 | 0.14 | 0.01 to 0.28 | .04* | 2.04 | .12 | 38 | .29 |
| rs878567 | |||||||||
| Overall symptoms | |||||||||
| CC vs G carriers | 4 | 743 | 0.02 | -0.14 to 0.17 | .85 | 0.19 | .81 | 0 | .883 |
| C carriers vs GG | 4 | 743 | 0.04 | -0.27 to 0.35 | .79 | 0.27 | .20 | 36 | .616 |
| C alleles vs G alleles | 4 | 1486 | 0.02 | -0.09 to 0.13 | .75 | 0.33 | .89 | 0 | .626 |
| Positive symptoms | |||||||||
| CC vs G carriers | 3 | 378 | -0.05 | -0.27 to 0.16 | .63 | 0.49 | .51 | 0 | .882 |
| C carriers vs GG | 3 | 378 | -0.12 | -0.48 to 0.24 | .51 | 0.66 | .79 | 0 | .658 |
| C alleles vs G alleles | 3 | 756 | -0.06 | -0.22 to 0.11 | .49 | 0.69 | .59 | 0 | .791 |
| Negative symptoms | |||||||||
| CC vs G carriers | 3 | 378 | 0.09 | -0.13 to 0.31 | .41 | 0.82 | .65 | 0 | .05 |
| C carriers vs GG | 3 | 378 | 0.08 | -0.51 to 0.68 | .78 | 0.28 | .10 | 57 | .931 |
| C alleles vs G alleles | 3 | 756 | 0.08 | -0.09 to 0.24 | .38 | 0.89 | .93 | 0 | .771 |
| rs1423691 | |||||||||
| Overall symptoms | |||||||||
| CC vs G carriers | 3 | 666 | -0.04 | -0.21 to 0.13 | .66 | 0.43 | .69 | 0 | .774 |
| C carriers vs GG | 3 | 666 | -0.05 | -0.46 to 0.36 | .80 | 0.26 | .07 | 62 | .419 |
| C alleles vs G alleles | 3 | 1332 | 0.00 | -0.11 to 0.12 | .97 | 0.04 | .86 | 0 | .825 |
| Positive symptoms | |||||||||
| CC vs G carriers | 2 | 302 | 0.06 | -0.30 to 0.18 | .63 | 0.48 | .44 | 0 | NA |
| C carriers vs GG | 2 | 302 | -0.16 | -0.55 to 0.23 | .42 | 0.81 | .62 | 0 | NA |
| C alleles vs G alleles | 2 | 604 | -0.07 | -0.26 to 0.11 | .45 | 0.76 | .49 | 0 | NA |
| Negative symptoms | |||||||||
| CC vs G carriers | 2 | 302 | 0.03 | -0.22 to 0.27 | .82 | 0.22 | .98 | 0 | NA |
| C carriers vs GG | 2 | 302 | -0.10 | -0.77 to 0.56 | .76 | 0.31 | .09 | 65 | NA |
| C alleles vs G alleles | 2 | 604 | 0.00 | -0.18 to 0.19 | .98 | 0.02 | .47 | 0 | NA |
Abbreviations: NA, not applicable; SMD, standardized mean difference.
*Significant difference (P < .05).
Figure 3.
Forest plot for the association with rs6295 polymorphism (C alleles vs G alleles) and improvement of positive symptoms.
Figure 4.
Forest plot for the association with rs6295 polymorphism (C alleles vs G alleles) and improvement of negative symptoms.
In regard to heterogeneity, a significant inter-study heterogeneity was observed only for negative symptoms data in the comparison between CG and G carriers (P=.03, I2 = 53%). No significant heterogeneity was detected in any of the other comparisons for rs6295 (Table 2).
rs878567 and Antipsychotic Drug Response
In the total population (4 studies, n=743), none of the comparisons revealed a significant correlation between the rs878567 polymorphism and changes in all 3 types of psychotic symptoms (overall symptoms, positive symptoms, and negative symptoms) (Table 2). There was no significant heterogeneity in any of these comparisons.
rs1423691 and Antipsychotic Drug Response
In the total population (3 studies, n = 666), none of the comparisons revealed a significant correlation between the rs1423691polymorphism and changes in all 3 types of psychotic symptoms (overall symptoms, positive symptoms, and negative symptoms) (Table 2). There was no significant heterogeneity in any of these comparisons.
Subgroup Analysis, Meta-Regression, and Publication Bias
In the subgroup analyses by race (Caucasian, Asian, and South Indian), no significant correlation was detected between any phenotype (change in overall symptoms, positive symptoms, or negative symptoms) and the rs6295 polymorphism (supplementary Table 1). No significant heterogeneity was found between subgroups (supplementary Table 1).
Meta-regression analysis of the rs6295 polymorphism indicated higher improvement in negative symptoms in G carriers vs CC carriers as the mean age increased (coefficient = -0.04, SE = 0.012, P=.02) (Table 3). This result may represent a potential source of the significant inter-study heterogeneity detected in the comparison between rs6295 CC and G carriers for negative symptom improvement. However, no impact of other moderator variables on the association between the rs6295 polymorphism and symptom improvement was found (Table 3).
Table 3.
Results of Meta-Regression Analyses for rs6295
| Moderator Variable | Coefficient | Standard Error | Test for Overall Effect | Heterogeneity | |
|---|---|---|---|---|---|
| P value | I2 (%) | ||||
| Overall symptoms | |||||
| CC vs G carriers | Study duration | 0.005 | 0.039 | .90 | 53 |
| Mean age | -0.028 | 0.014 | .08 | 33.76 | |
| Male ratio | 0.009 | 0.009 | .34 | 51.02 | |
| C carriers vs GG | Study duration | 0.008 | 0.027 | .77 | 0 |
| Mean age | -0.015 | 0.015 | .37 | 0 | |
| Male ratio | -0.006 | 0.009 | .56 | 0 | |
| C alleles vs G alleles | Study duration | 0.003 | 0.021 | .91 | 36.44 |
| Mean age | -0.015 | 0.015 | .37 | 0 | |
| Male ratio | 0.003 | 0.006 | .62 | 35.5 | |
| Positive symptoms | |||||
| CC vs G carriers | Study duration | -0.022 | 0.030 | .49 | 0 |
| Mean age | -0.026 | 0.012 | .08 | 0 | |
| Male ratio | 0.003 | 0.007 | .65 | 0 | |
| C carriers vs GG | Study duration | 0.008 | 0.036 | .84 | 0 |
| Mean age | -0.010 | 0.021 | .65 | 0 | |
| Male ratio | -0.008 | 0.010 | .48 | 0 | |
| C alleles vs G alleles | Study duration | -0.010 | 0.020 | .63 | 0 |
| Mean age | -0.015 | 0.010 | .17 | 0 | |
| Male ratio | 0.000 | 0.005 | .98 | 0 | |
| Negative symptoms | |||||
| CC vs G carriers | Study duration | 0.031 | 0.043 | .49 | 58.11 |
| Mean age | -0.040 | 0.012 | .02* | 0 | |
| Male ratio | 0.009 | 0.010 | .40 | 57.72 | |
| C carriers vs GG | Study duration | -0.001 | 0.038 | .98 | 0 |
| Mean age | -0.014 | 0.021 | .54 | 0 | |
| Male ratio | -0.002 | 0.010 | .82 | 0 | |
| C alleles vs G alleles | Study duration | 0.015 | 0.027 | .60 | 45.44 |
| Mean age | -0.023 | 0.010 | .06 | 0 | |
| Male ratio | 0.004 | 0.007 | .54 | 45.69 | |
*Significant difference (P < .05).
Visual inspection of funnel-plots for each distribution of the 3 HTR1A polymorphisms revealed no evidence of publication bias (supplementary Figure 1). Egger’s test for publication bias did not suggest evidence of publication bias for any of the performed comparisons for the 3 HTR1A polymorphisms (supplementary Figure 1; Table 2).
Discussion
To our knowledge, this is the first meta-analysis investigating the impact of HTR1A polymorphisms (rs6295, rs1423691, rs878567) on the efficacy of antipsychotic medications in patients with schizophrenia. The analyses were performed employing 3 efficacy parameters, that is, improvement in overall symptoms, positive symptoms, and negative symptoms; 10 studies and 1281 patients were included. Our results indicate that the investigated HTR1A polymorphisms are unlikely to impact overall symptom or positive symptom change during antipsychotic treatment. However, the rs6295 polymorphism showed a small effect on negative symptom improvement in the comparison C vs G allele (P=.04, SMD = 0.14, CI = 0.01 to 0.28).
5-HT1AR is considered to play an important role in antipsychotic mechanisms of action both directly and through indirect mechanisms (Meltzer and Massey, 2011; Newman-Tancredi and Kleven, 2011). As a “direct” mechanism, some antipsychotic drugs are known to improve psychiatric symptoms via a partial agonistic action exerted on 5-HT1AR (Jordan et al., 2002; Odagaki and Toyoshima, 2007; Murasaki et al., 2008; Fagiolini et al., 2010; Kishi et al., 2013a; Lerond et al., 2013). As an “indirect” effect, second-generation antipsychotics are known to indirectly cause dopamine release in the prefrontal cortex via postsynaptic 5-HT1AR and 5-HT1A autoreceptors present in the raphe nucleus (Di Matteo et al., 2000; Ichikawa et al., 2001; Diaz-Mataix et al., 2005; Bortolozzi et al., 2010; Celada et al., 2013; Huang et al., 2014). Functional SNPs in the HTR1A gene have a relevant impact on 5-HT1AR level/activity. Particularly, rs6295 in the promoter region of HTR1A is known to be a functional polymorphism (Jacobsen et al., 2008; Albert and Fiori, 2014). In raphe cells, the C-G change impairs the binding of nuclear proteins (eg, Deaf1) to a palindrome DNA element located at this polymorphism (Lemonde et al., 2003; Albert and Fiori, 2014). The rs6295 G allele is associated with a reduced efficiency of Deaf-1 binding, resulting in increased 5-HT1A autoreceptor expression in the raphe nucleus, where serotonin neurons originate (Albert, 2012). In contrast, 5-HT1AR expression is hypothesized to be decreased on the postsynaptic side, such as in the prefrontal cortex (Czesak et al., 2006). Consistently, Deaf1 -/- mice show a 50% increase in 5-HT1A RNA in the raphe, but a 30% decrease in the prefrontal cortex, confirming the role of Deaf1 in regulating HTR1A gene expression (Czesak et al., 2012). Basic study data suggested that lowered postsynaptic 5-HT1AR density may lead to reduced serotonergic signal transduction, which may in turn reduce dopamine release in the prefrontal cortex (Varrault et al., 1992; Bortolozzi et al., 2010). These findings are consistent with the results of the present meta-analysis that support the hypothesis of rs6295 selective impact on negative symptoms. However, this result should be interpreted with caution given that the effect was small (SMD = 0.14).
The other polymorphisms investigated by the present meta-analysis, that is, rs878567 and rs1423691, are located in the HTR1A downstream region, and they involve no amino acid substitutions (Gonzalez-Castro et al., 2013). It has been suggested that the rs878567 polymorphism might be related to mood disorders (particularly major depressive disorder) (Kishi et al., 2013b), methamphetamine-induced psychosis (Kishi et al., 2010), and attention deficit-hyperactivity disorder (Park et al., 2013) and that rs1423691 may also be associated with attention deficit-hyperactivity disorder (Park et al., 2013). However, there are no reports supporting a functional effect of these polymorphisms, and the present meta-analysis did not reveal significant correlations between these SNPs and clinical responses to antipsychotics. A limited number of studies was available for these 2 polymorphisms, and further data would be helpful in clarifying their role in antipsychotic response.
The limitations of the present meta-analysis should be considered. First, the number of studies included was small, particularly for rs878567 and rs1423691. Second, only candidate gene studies and not genome-wide association studies were included. Third, nongenetic stratification factors may have influenced the results. Indeed, studies using different antipsychotic drugs were included, and 5-HT1AR binding affinity is not the same for all antipsychotics. It was not possible to perform stratified analyses to account for this source of heterogeneity, since the antipsychotic drugs used varied greatly among the studies. Attention should also be paid to other sources of stratification among the included studies, such as ethnicity, diagnosis, and other clinical differences (eg, disease duration), duration of the study, and tools used to assess symptom improvement.
In conclusion, the present meta-analysis indicated that antipsychotic treatment may be more effective in improving negative symptoms in C allele than in G allele carriers of the HTR1A rs6295 polymorphism. This finding is consistent with data obtained from basic research on the importance of 5-HT1AR in antipsychotic mechanisms of action and the role of the HTR1A rs6295 polymorphism in regulating 5-HT1AR expression. However, this finding showed a small effect size, and relatively few studies and patients were included in the present meta-analysis; thus, the result should be confirmed in larger samples possibly including clinically homogeneous patients.
Statement of Interest
Dr. Takekita has received grant funding from Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) and speaker’s honoraria from Dainippon-Sumitomo Pharma, Otsuka, Meiji-Seika Pharma, Janssen Pharmaceutical, and Ono Pharmaceutical within the past 3 years. Dr. Kato has received grant funding from Grant-in-Aid for Scientific Research (C) from JSPS and speaker’s honoraria from Dainippon-Sumitomo Pharma, Otsuka, Meiji-Seika Pharma, Eli Lilly, MSD K.K., GlaxoSmithkline, Pfizer, Shionogi, and Ono Pharmaceutical within the past 3 years. Mr. Koshikawa is or has been consultant/speaker for Dainippon-Sumitomo Pharma and Janssen Pharmaceutical. Dr. Tajika has received honoraria for speaking at a meeting sponsored by Eli Lilly and Tanabe-Mitsubishi. Dr. Kinoshita has received grant/research support or honoraria from and been a speaker for Dainippon-Sumitomo Pharma, Otsuka, Meiji-Seika Pharma, Janssen Pharmaceutical, Daiichi-Sankyo company, Takeda Pharmaceutical, Eli Lilly, MSD K.K. Shionogi, Astellas Pharma, Eisai, GlaxoSmithkline, and Ono Pharmaceutical. Dr. Serretti is or has been consultant/speaker for Abbott, Abbvie, Angelini, Astra Zeneca, Clinical Data, Boheringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, Innovapharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polipharma, Sanofi, and Servier. The other authors declare no competing interests. All authors declare that they have no direct conflicts of interest relevant to this study.
Supplementary Material
Acknowledgments
We thank Dr. Gavin P. Reynolds, Dr. Ritushree Kukreti, Dr. Masashi Ikeda, Dr. Marta Bosia and Dr. Tomiki Sumiyoshi, for providing data.
Dr. Takekita had access to all study data and takes full responsibility for its integrity and accuracy of the analysis. Drs. Takekita, Fabbri, Koshikawa, and Tajika participated in study conception and design, data acquisition, and statistical analysis. The manuscript was written by Drs. Takekita and Fabbri. Drs. Kato, Kinoshita, and Serretti supervised the review.
References
- Albert PR. (2012) Transcriptional regulation of the 5-HT1A receptor: implications for mental illness. Philos Trans R Soc Lond B Biol Sci 367:2402–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert PR, Lemonde S. (2004) 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist 10:575–593. [DOI] [PubMed] [Google Scholar]
- Albert PR, Fiori LM. (2014) Transcriptional dys-regulation in anxiety and major depression: 5-HT1A gene promoter architecture as a therapeutic opportunity. Curr Pharm Des 20:3738–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar S, Qian Z, Shah R, Rahbek B, Knudsen GM. (2003) The 5-HT1A serotonin receptor is located on calbindin- and parvalbumin-containing neurons in the rat brain. Brain Res 959:58–67. [DOI] [PubMed] [Google Scholar]
- Barnes NM, Sharp T. (1999) A review of central 5-HT receptors and their function. Neuropharmacology 38:1083–1152. [DOI] [PubMed] [Google Scholar]
- Bortolozzi A, Masana M, Diaz-Mataix L, Cortes R, Scorza MC, Gingrich JA, Toth M, Artigas F. (2010) Dopamine release induced by atypical antipsychotics in prefrontal cortex requires 5-HT(1A) receptors but not 5-HT(2A) receptors. Int J Neuropsychopharmacol 13:1299–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosia M, Lorenzi C, Pirovano A, Guglielmino C, Cocchi F, Spangaro M, Bramanti P, Smeraldi E, Cavallaro R. (2015) COMT Val158Met and 5-HT1A-R -1019 C/G polymorphisms: effects on the negative symptom response to clozapine. Pharmacogenomics 16:35–44. [DOI] [PubMed] [Google Scholar]
- Burnet PW, Eastwood SL, Harrison PJ. (1996) 5-HT1A and 5-HT2A receptor mRNAs and binding site densities are differentially altered in schizophrenia. Neuropsychopharmacology 15:442–455. [DOI] [PubMed] [Google Scholar]
- Celada P, Puig MV, Artigas F. (2013) Serotonin modulation of cortical neurons and networks. Front Integr Neurosci 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa A, Lia L, Han C, Lee SJ, Pae CU, Serretti A. (2013) Investigation of epistasis between DAOA and 5HTR1A variants on clinical outcomes in patients with schizophrenia. Genet Test Mol Biomarkers 17:504–507. [DOI] [PubMed] [Google Scholar]
- Crisafulli C, Chiesa A, Han C, Lee SJ, Park MH, Balzarro B, Andrisano C, Patkar AA, Pae CU, Serretti A. (2012) Case-control association study for 10 genes in patients with schizophrenia: influence of 5HTR1A variation rs10042486 on schizophrenia and response to antipsychotics. Eur Arch Psychiatry Clin Neurosci 262:199–205. [DOI] [PubMed] [Google Scholar]
- Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR. (2006) Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J Neurosci 26:1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M, Le Francois B, Millar AM, Deria M, Daigle M, Visvader JE, Anisman H, Albert PR. (2012) Increased serotonin-1A (5-HT1A) autoreceptor expression and reduced raphe serotonin levels in deformed epidermal autoregulatory factor-1 (Deaf-1) gene knock-out mice. J Biol Chem 287:6615–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188. [DOI] [PubMed] [Google Scholar]
- Di Matteo V, Di Giovanni G, Di Mascio M, Esposito E. (2000) Biochemical and electrophysiological evidence that RO 60–0175 inhibits mesolimbic dopaminergic function through serotonin(2C) receptors. Brain Res 865:85–90. [DOI] [PubMed] [Google Scholar]
- Diaz-Mataix L, Scorza MC, Bortolozzi A, Toth M, Celada P, Artigas F. (2005) Involvement of 5-HT1A receptors in prefrontal cortex in the modulation of dopaminergic activity: role in atypical antipsychotic action. J Neurosci 25:10831–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago A, Giegling I, Schafer M, Hartmann AM, Friedl M, Konte B, Moller HJ, De Ronchi D, Stassen HH, Serretti A, Rujescu D. (2013) AKAP13, CACNA1, GRIK4 and GRIA1 genetic variations may be associated with haloperidol efficacy during acute treatment. Eur Neuropsychopharmacol 23:887–894. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiolini A, Canas F, Gallhofer B, Larmo I, Levy P, Montes JM, Papageorgiou G, Zink M, Rossi A. (2010) Strategies for successful clinical management of schizophrenia with ziprasidone. Expert Opin Pharmacother 11:2199–2220. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Castro TB, Tovilla-Zarate CA, Juarez-Rojop I, Pool Garcia S, Genis A, Nicolini H, Lopez Narvaez L. (2013) Association of 5HTR1A gene variants with suicidal behavior: case-control study and updated meta-analysis. J Psychiatr Res 47:1665–1672. [DOI] [PubMed] [Google Scholar]
- Gu H, Liu C, Liu C, Chen M, Zhang Q, Zhai J, Wang K, Ji F, Xu Z, Shen Q, Bao X, Chen X, Li J, Dong Q, Chen C. (2013) The combined effects of the 5- HTTLPR and HTR1A rs6295 polymorphisms modulate decision making in schizophrenia patients. Genes Brain Behav 12:133–139. [DOI] [PubMed] [Google Scholar]
- Gupta M, Jain S, Moily NS, Kaur H, Jajodia A, Purushottam M, Kukreti R. (2012) Genetic studies indicate a potential target 5-HTR(3B) for drug therapy in schizophrenia patients. Am J Med Genet B Neuropsychiatr Genet 159B:1006–1008. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Nishino N, Nakai H, Tanaka C. (1991) Increase in serotonin 5-HT1A receptors in prefrontal and temporal cortices of brains from patients with chronic schizophrenia. Life Sci 48:355–363. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Panos JJ, Kwon S, Oyamada Y, Rajagopal L, Meltzer HY. (2014) Comparative effect of lurasidone and blonanserin on cortical glutamate, dopamine, and acetylcholine efflux: role of relative serotonin (5-HT)2A and DA D2 antagonism and 5-HT1A partial agonism. J Neurochem 128:938–949. [DOI] [PubMed] [Google Scholar]
- Ichikawa J, Ishii H, Bonaccorso S, Fowler WL, O’Laughlin IA, Meltzer HY. (2001) 5-HT(2A) and D(2) receptor blockade increases cortical DA release via 5-HT(1A) receptor activation: a possible mechanism of atypical antipsychotic-induced cortical dopamine release. J Neurochem 76:1521–1531. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yamanouchi Y, Kinoshita Y, Kitajima T, Yoshimura R, Hashimoto S, O’Donovan MC, Nakamura J, Ozaki N, Iwata N. (2008) Variants of dopamine and serotonin candidate genes as predictors of response to risperidone treatment in first-episode schizophrenia. Pharmacogenomics 9:1437–1443. [DOI] [PubMed] [Google Scholar]
- Jacobsen KX, Vanderluit JL, Slack RS, Albert PR. (2008) HES1 regulates 5-HT1A receptor gene transcription at a functional polymorphism: essential role in developmental expression. Mol Cell Neurosci 38:349–358. [DOI] [PubMed] [Google Scholar]
- Jordan S, Koprivica V, Chen R, Tottori K, Kikuchi T, Altar CA. (2002) The antipsychotic aripiprazole is a potent, partial agonist at the human 5-HT1A receptor. Eur J Pharmacol 441:137–140. [DOI] [PubMed] [Google Scholar]
- Kato M, Serretti A. (2010) Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 15:473–500. [DOI] [PubMed] [Google Scholar]
- Kim YK, Yoon HK. (2011) Effect of serotonin-related gene polymorphisms on pathogenesis and treatment response in Korean schizophrenic patients. Behav Genet 41:709–715. [DOI] [PubMed] [Google Scholar]
- Kishi T, Tsunoka T, Ikeda M, Kitajima T, Kawashima K, Okochi T, Okumura T, Yamanouchi Y, Kinoshita Y, Ujike H, Inada T, Yamada M, Uchimura N, Sora I, Iyo M, Ozaki N, Iwata N. (2010) Serotonin 1A receptor gene is associated with Japanese methamphetamine-induced psychosis patients. Neuropharmacology 58:452–456. [DOI] [PubMed] [Google Scholar]
- Kishi T, Meltzer HY, Iwata N. (2013a) Augmentation of antipsychotic drug action by azapirone 5-HT1A receptor partial agonists: a meta-analysis. Int J Neuropsychopharmacol 16:1259–1266. [DOI] [PubMed] [Google Scholar]
- Kishi T, Yoshimura R, Fukuo Y, Okochi T, Matsunaga S, Umene-Nakano W, Nakamura J, Serretti A, Correll CU, Kane JM, Iwata N. (2013b) The serotonin 1A receptor gene confer susceptibility to mood disorders: results from an extended meta-analysis of patients with major depression and bipolar disorder. Eur Arch Psychiatry Clin Neurosci 263:105–118. [DOI] [PubMed] [Google Scholar]
- Lane HY, Liu YC, Huang CL, Chang YC, Wu PL, Lu CT, Chang WH. (2006) Risperidone-related weight gain: genetic and nongenetic predictors. J Clin Psychopharmacol 26:128–134. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, Ou XM, Albert PR. (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci 23:8788–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerond J, Lothe A, Ryvlin P, Bouvard S, d’Amato T, Ciumas C, Dalery J, Poulet E, Saoud M. (2013) Effects of aripiprazole, risperidone, and olanzapine on 5-HT1A receptors in patients with schizophrenia. J Clin Psychopharmacol 33:84–89. [DOI] [PubMed] [Google Scholar]
- Masellis M, Basile VS, Meltzer HY, Lieberman JA, Sevy S, Goldman DA, Hamblin MW, Macciardi FM, Kennedy JL. (2001) Lack of association between the T-->C 267 serotonin 5-HT6 receptor gene (HTR6) polymorphism and prediction of response to clozapine in schizophrenia. Schizophr Res 47:49–58. [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Massey BW. (2011) The role of serotonin receptors in the action of atypical antipsychotic drugs. Curr Opin Pharmacol 11:59–67. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner R, Schuhmacher A, Kuhn KU, Cvetanovska G, Rujescu D, Zill P, Quednow BB, Rietschel M, Wolwer W, Gaebel W, Wagner M, Maier W. (2009) Functional serotonin 1A receptor variant influences treatment response to atypical antipsychotics in schizophrenia. Pharmacogenet Genomics 19:91–94. [DOI] [PubMed] [Google Scholar]
- Murasaki M, Nishikawa H, Ishibashi T. (2008) Dopamine-serotonin antagonist: Receptor binding profile of a novel antipsychotic blonanserin. Japanese Clinical psychopharmacology 11:845–854. [Google Scholar]
- Newman-Tancredi A, Kleven MS. (2011) Comparative pharmacology of antipsychotics possessing combined dopamine D2 and serotonin 5-HT1A receptor properties. Psychopharmacology (Berl) 216:451–473. [DOI] [PubMed] [Google Scholar]
- Niitsu T, Fabbri C, Bentini F, Serretti A. (2013) Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry 45:183–194. [DOI] [PubMed] [Google Scholar]
- Odagaki Y, Toyoshima R. (2007) 5-HT1A receptor agonist properties of antipsychotics determined by [35S]GTPgammaS binding in rat hippocampal membranes. Clin Exp Pharmacol P 34:462–466. [DOI] [PubMed] [Google Scholar]
- Park YH, Lee KK, Kwon HJ, Ha M, Kim EJ, Yoo SJ, Paik KC, Lim MH. (2013) Association between HTR1A gene polymorphisms and attention deficit hyperactivity disorder in Korean children. Genet Test Mol Biomarkers 17:178–182. [DOI] [PubMed] [Google Scholar]
- Reynolds GP, Arranz B, Templeman LA, Fertuzinhos S, San L. (2006) Effect of 5-HT1A receptor gene polymorphism on negative and depressive symptom response to antipsychotic treatment of drug-naive psychotic patients. Am J Psychiatry 163:1826–1829. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi T, Stockmeier CA, Overholser JC, Dilley GE, Meltzer HY. (1996) Serotonin1A receptors are increased in postmortem prefrontal cortex in schizophrenia. Brain Res 708:209–214. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi T, Tsunoda M, Higuchi Y, Itoh T, Seo T, Itoh H, Suzuki M, Kurachi M. (2010) Serotonin-1A receptor gene polymorphism and the ability of antipsychotic drugs to improve attention in schizophrenia. Adv Ther 27:307–313. [DOI] [PubMed] [Google Scholar]
- Takekita Y, Fabbri C, Kato M, Nonen S, Sakai S, Sunada N, Koshikawa Y, Wakeno M, Okugawa G, Kinoshita T, Serretti A. (2015) HTR1A gene polymorphisms and 5-HT1A receptor partial agonist antipsychotics efficacy in schizophrenia. J Clin Psychopharmacol 35:220–227. [DOI] [PubMed] [Google Scholar]
- Tang H, Dalton CF, Srisawat U, Zhang ZJ, Reynolds GP. (2014) Methylation at a transcription factor-binding site on the 5-HT1A receptor gene correlates with negative symptom treatment response in first episode schizophrenia. Int J Neuropsychopharmacol 17:645–649. [DOI] [PubMed] [Google Scholar]
- Tauscher J, Kapur S, Verhoeff NP, Hussey DF, Daskalakis ZJ, Tauscher-Wisniewski S, Wilson AA, Houle S, Kasper S, Zipursky RB. (2002) Brain serotonin 5-HT(1A) receptor binding in schizophrenia measured by positron emission tomography and [11C]WAY-100635. Arch Gen Psychiatry 59:514–520. [DOI] [PubMed] [Google Scholar]
- Terzic T, Kastelic M, Dolzan V, Plesnicar BK. (2015) Influence of 5-HT1A and 5-HTTLPR genetic variants on the schizophrenia symptoms and occurrence of treatment-resistant schizophrenia. Neuropsychiatr Dis Treat 11:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cochrane Collaboration (2011) Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0. In. [Google Scholar]
- Varnas K, Halldin C, Hall H. (2004) Autoradiographic distribution of serotonin transporters and receptor subtypes in human brain. Hum Brain Mapp 22:246–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Journot L, Audigier Y, Bockaert J. (1992) Transfection of human 5-hydroxytryptamine1A receptors in NIH-3T3 fibroblasts: effects of increasing receptor density on the coupling of 5-hydroxytryptamine1A receptors to adenylyl cyclase. Mol Pharmacol 41:999–1007. [PubMed] [Google Scholar]
- Wang D, Fu DJ, Wu X, Shapiro A, Favis R, Savitz A, Chung H, Alphs L, Gopal S, Haas M, Cohen N, Li Q. (2015) Large-scale candidate gene study to identify genetic risk factors predictive of paliperidone treatment response in patients with schizophrenia. Pharmacogenet Genomics 25:173–185. [DOI] [PubMed] [Google Scholar]
- Wang L, Fang C, Zhang A, Du J, Yu L, Ma J, Feng G, Xing Q, He L. (2008) The --1019 C/G polymorphism of the 5-HT(1)A receptor gene is associated with negative symptom response to risperidone treatment in schizophrenia patients. J Psychopharmacol 22:904–909. [DOI] [PubMed] [Google Scholar]
- Wedzony K, Mackowiak M, Czyrak A, Fijal K, Michalska B. (1997) Single doses of MK-801, a non-competitive antagonist of NMDA receptors, increase the number of 5-HT1A serotonin receptors in the rat brain. Brain Res 756:84–91. [DOI] [PubMed] [Google Scholar]
- Wu S, Comings DE. (1999) A common C-1018G polymorphism in the human 5-HT1A receptor gene. Psychiatr Genet 9:105–106. [DOI] [PubMed] [Google Scholar]
- Zhao X, Huang Y, Li J, Ma H, Jin Q, Wang Y, Wu L, Zhu G. (2012) Association between the 5-HT1A receptor gene polymorphism (rs6295) and antidepressants: a meta-analysis. Int Clin Psychopharmacol 27:314–320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.