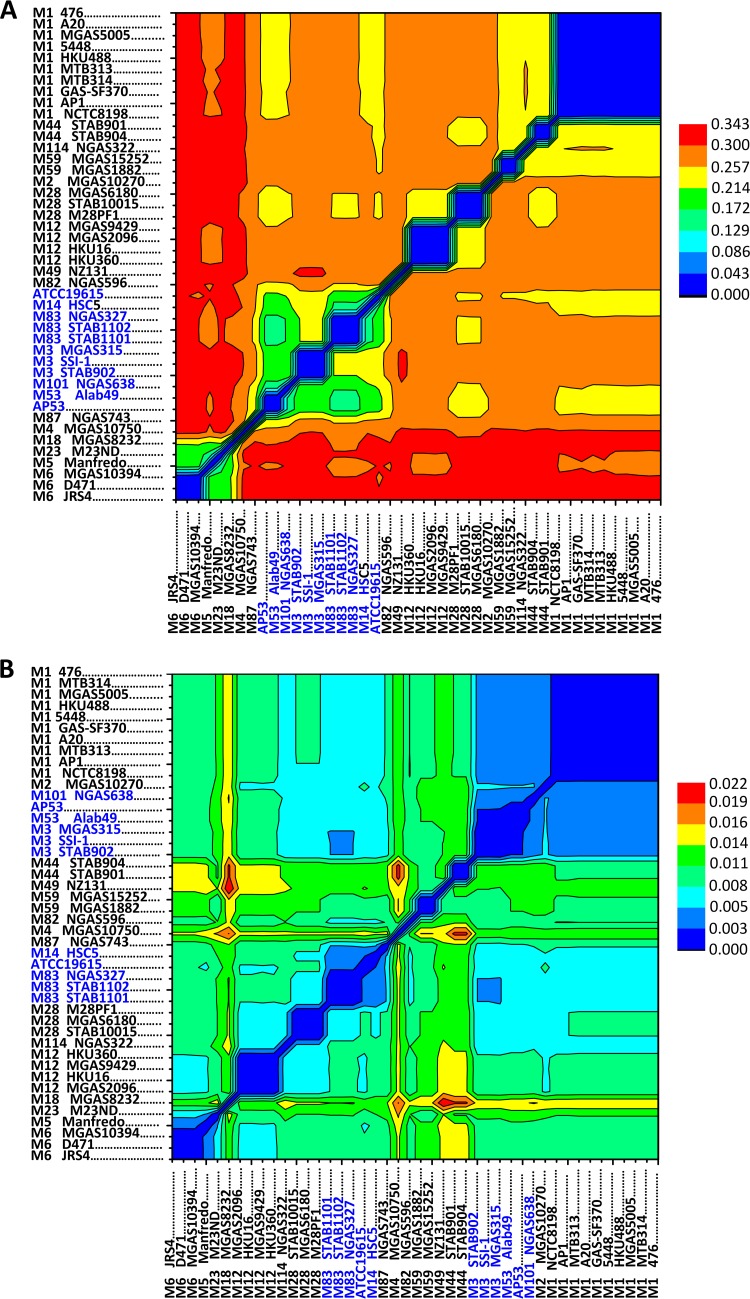
FIG 3.
GAS strain clustering based on pairwise distances of all GAS genomes measured by base substitutions per site. (A) The substitution rate was determined for the core genome of GAS, containing 71,558 SNP loci. The skin-tropic strains, i.e., AP53, M53 Alab49, M101 NGAS638, M83 strains, M3 strains, ATCC 19615, and M14 HSC5, are clustered together and highlighted in blue. (B) The substitution rates for seven housekeeping genes, viz., gki, gtr, murI, mutS, recP, xpt, and yqiL, were determined, generating 434 SNP loci. The strains clustered in panel A are subdivided into distinct clusters in panel B. The pairwise distance was estimated based on the model of maximum composite likelihood. The clustering relationship was inferred by using the maximum likelihood method with a bootstrap value of 1,000.