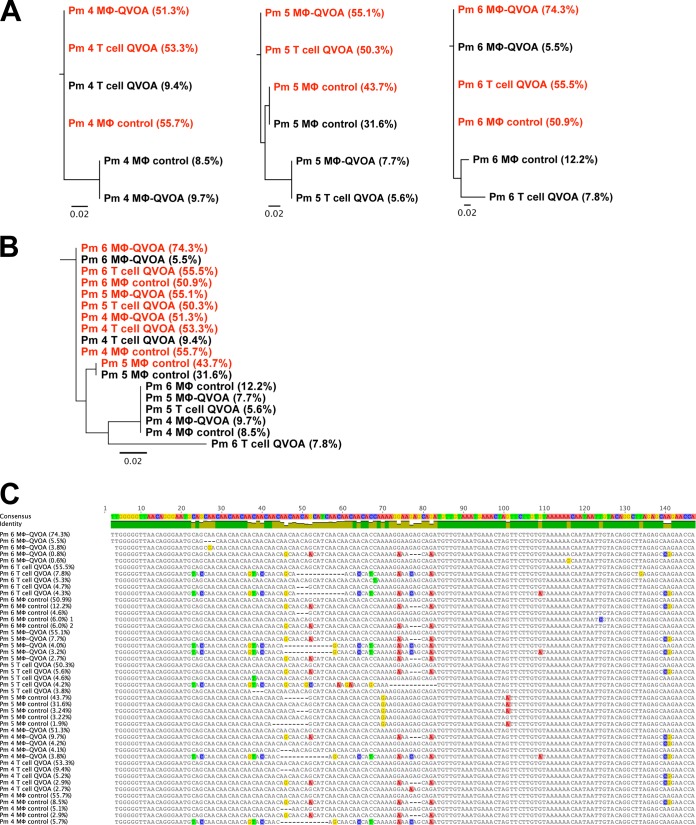
FIG 7.
Sequence analyses of virus produced in CD4+ T cell and macrophage QVOAs. Supernatant was collected from the spleen Mϕ-QVOA and spleen T cell QVOA for Pm4, Pm5, and Pm6. Viral RNA was isolated, and a two-round nested PCR for SIV env was performed. CD11b+ macrophage wells without CEMx174 cells from the Mϕ-QVOA were used as controls. The most frequent (red) and the second most frequent (black) sequences are depicted along with the frequency of the viral clone (indicated in parentheses). (A and B) A tree of the nucleotide sequence alignment for each animal (A) and a phylogenetic tree of all clones (B) are shown. (C) Comparison of the nucleotide sequences of the prevailing clones with the consensus sequence, with percentages signifying the frequency of the clones. The scale bar represents the distance between the sequences. Analyses were performed by Geneious (version 8.0) software.