ABSTRACT
We provide here, for the first time, insights into the initial infection stages of a large spindle-shaped archaeal virus and explore the following life cycle events. Our observations suggest that Sulfolobus monocaudavirus 1 (SMV1) exhibits a high adsorption rate and that virions adsorb to the host cells via three distinct attachment modes: nosecone association, body association, and body/tail association. In the body/tail association mode, the entire virion, including the tail(s), aligns to the host cell surface and the main body is greatly flattened, suggesting a possible fusion entry mechanism. Upon infection, the intracellular replication cycle lasts about 8 h, at which point the virions are released as spindle-shaped tailless particles. Replication of the virus retarded host growth but did not cause lysis of the host cells. Once released from the host and at temperatures resembling that of its natural habitat, SMV1 starts developing one or two tails. This exceptional property of undergoing a major morphological development outside, and independently of, the host cell has been reported only once before for the related Acidianus two-tailed virus. Here, we show that SMV1 can develop tails of more than 900 nm in length, more than quadrupling the total virion length.
IMPORTANCE Very little is known about the initial life cycle stages of viruses infecting hosts of the third domain of life, Archaea. This work describes the first example of an archaeal virus employing three distinct association modes. The virus under study, Sulfolobus monocaudavirus 1, is a representative of the large spindle-shaped viruses that are frequently found in acidic hot springs. The results described here will add valuable knowledge about Archaea, the least studied domain in the virology field.
INTRODUCTION
Archaeal viruses constitute an integral part of the virosphere, and they are a ubiquitous feature of archaeal existence. Many Archaea are recognized as extremophiles, inhabiting extreme environments such as hot springs and solar salterns in high abundance (1, 2). Thus, viruses infecting extremophilic Archaea can be considered key players in the complex population dynamics in these environments. Through host infection, viruses can influence microbial diversity by introducing genetic variation, affect host cell physiology, and directly kill their hosts by cell lysis (3, 4). However, we have only a rudimentary understanding of archaeal virus-host interactions. Much of the limited knowledge that we do have comes from studying the virus-host interplay in Sulfolobales species. Of the about 100 isolated archaeal viruses, more than 30% infect hyperthermophilic Sulfolobales hosts (5–7). Among these viruses, distinct characteristics are found: unique bottle, droplet, and spindle shapes; extracellular virion development; and unique proteins with unknown functions (8). These distinctive characteristics are likely to influence the interplay with their hosts and give rise to unique life cycle traits. This has proven true for the rod-shaped Sulfolobus virus SIRV2, which has been shown to have an exceptional egress mechanism involving pyramid-like structures. To date, SIRV2 remains the most well characterized archaeal virus, and very little is known about the initial entry processes and later egress mechanisms of other archaeal viruses (9–11).
As a group, viruses with spindle-shaped virions, single or two tailed, are common in and exclusive to the Archaea (12). Despite this architecture being the most common found in Archaea-dominated habitats, studies of these viruses have mostly been confined to biochemical and genetic characterizations of their virions (13–16). Relatively little is known about the relationships between them and their hosts; in particular, insights into the entry process are lacking. Recently, it was suggested to group five of the large spindle-shaped viruses together into a new superfamily based on structural similarities and a shared set of core genes (15). The group comprises Acidianus two-tailed virus (ATV), Sulfolobus tengchongensis spindle-shaped virus 1 (STSV1) and STSV2, Acidianus tailed spindle-shaped virus (ATSV), and Sulfolobus monocaudavirus 1 (SMV1) (14–18). The virion structures of these large spindle-shaped viruses are often pleomorphic, and their tails also vary greatly in length. For example, ATSV tails range in length from 35 to 720 nm, and ATV has been observed to develop two elongated tails once released from the host (15, 19). In order to elucidate the life cycles of these fascinating viruses, a robust model system is needed.
SMV1 was originally isolated from an acidic high-temperature hot spring in Yellowstone National Park (17). Virions of SMV1 are spindle shaped (averaging 200 by 70 nm) with a single tail varying in length from 20 to 500 nm and a nose-like structure on the opposite pole, which occasionally extends to generate a second tail. SMV1 has a genome size of 48.8 kb with 51 putative open reading frames (ORFs) (one major coat protein is predicted). On infection in Sulfolobus islandicus Rey15A, growth retardation occurs, but no evidence for cell lysis has been observed, and no clear plaques have been seen on Gelrite plates (17). It has proven easy to reproduce SMV1 in S. islandicus to obtain high virus titers. Thus, this virus-host system has been well established in our lab and represents a valuable model to study the virus-host interactions of large spindle-shaped viruses. Until now, the entry mechanisms of these viruses have not been investigated. Here, we studied the life cycle of SMV1 when infecting S. islandicus ΔC1C2 (13), with special focus on the early stages of infection.
MATERIALS AND METHODS
Virus propagation and purification.
SMV1 was propagated in S. islandicus ΔC1C2 (20). The host culture was grown in Sulfolobus medium supplemented with 0.2% (wt/vol) tryptone, 0.1% (wt/vol) yeast extract, 0.2% (wt/vol) sucrose, and 0.002% (wt/vol) uracil (TYS+U medium) (21). Cultures were started from −80°C stock; cells were transferred to 50 ml TYS+U medium and incubated at 78°C. After 24 h of propagation, the 50-ml cell culture was transferred to 950 ml of preheated (78°C) TYS+U medium. The culture was grown to an optical density at 600 nm (OD600) of 0.2 to 0.3 (typically 24 h), at which time point the host culture was infected with SMV1 stock. The supernatant containing the virus particles was collected at 48 to 72 h postinfection (hpi) and concentrated by ultrafiltration using 1,000,000-molecular-weight-cutoff (MWCO) centrifugal filter units (Sartorius, Aubagne Cedex, France). The virus fraction was washed two times with 10 ml 10 mM Tris-acetate buffer (pH 6) to exchange the medium with storage buffer. The virus titer was determined by plaque assay as described below. Virus samples were stored at 4°C until used.
Susceptibility of Sulfolobus strains to SMV1.
Five Sulfolobus strains, i.e., S. islandicus LAL14/1 (22), S. islandicus HVE10/4 (23), S. islandicus REY 15A (23), S. islandicus ΔC1C2 (20), and S. solfataricus 5E6 (11), were tested for susceptibility to SMV1. All strains were cultivated in TYS+U medium at 78°C with aeration at 150 rpm. Six 50-ml flasks of each culture were set up; three were uninfected controls, and three were infected with a multiplicity of infection (MOI) of 5. One milliliter was taken from each culture every 3 h for 48 h, and OD600 values were measured. To investigate population dynamics, identical growth experiments were set up with S. islandicus ΔC1C2 using an MOI range of 0.01 to 5. The cultures were monitored for 48 h.
One-step growth curve and plaque assay.
Plaque assays were performed in order to establish a one-step growth curve. Samples of cell-free supernatant from the time course infection at an MOI of 0.1 (see above) were serially diluted, and 10 μl of each diluted sample was mixed with 2 ml preheated fresh S. islandicus ΔC1C2 cells, followed by 2 ml preheated 0.4% (wt/vol) Phytagel (Sigma-Aldrich, USA). The mixture was layered over a 0.7% (wt/vol) solid Phytagel layer. Plates were incubated for 2 days at 78°C. Single plaques were counted, and PFU were determined.
Virus stability.
All virus stability experiments were carried out using highly purified virus stocks. Treatments included (i) detergents, (ii) proteinase K, (iii) different temperatures, and (iv) UV radiation. Incubations were carried out in either Tris-acetate buffer (pH 6) or treatment solution. After each treatment, infectivity was determined using the plaque assay, and virus particles were visualized by negative-staining transmission electron microscopy. Briefly, virus particles were diluted 10-fold in the following treatment solutions: Triton X-100 (0.1% or 0.01% [wt/vol]), proteinase K (0.2 mg/ml), or Tris-acetate buffer (pH 6). The temperature conditions for the individual dilutions are listed in Table S1 in the supplemental material. To generate UV-inactivated virus stocks, virus particles diluted 10-fold in Tris-acetate buffer (pH 6) were irradiated in open petri dishes with either 40 mJ/cm2 or 1 J/cm2 of 254-nm UV light using a UV Stratalinker 1800 (Stratagene, La Jolla, CA).
Adsorption assay.
For the adsorption assay, S. islandicus ΔC1C2 cells (OD600 = 0.2; 108 cells/ml) were infected using an MOI of 0.1. At defined time intervals, a sample of infected culture was removed and the adsorption stopped by immediate centrifugation (10,000 × g, 5 min, room temperature [RT]). The number of remaining PFU was determined by plaque assay and compared to the number of PFU present in a cell-free control mixture incubated at 78°C. The adsorption rate constant (k) was calculated using the formula, k = 2.3/Bt × log10(P0/P) (24), where P0 = PFU per milliliter at zero time, P = virus not adsorbed at time t min, and B = concentration of host cells as number of cells per milliliter (24).
Membrane integrity analysis.
The membrane integrity of host cells was assessed using the Live/Dead BacLight bacterial viability kit (Molecular Probes, OR, USA) with minor modifications to the manufacturer's protocol. Briefly, 500-μl samples were taken at regular intervals from the infected (MOI of 3) and control cultures, and cells were collected by centrifugation (1,000 × g, 20 min, RT). The cells were resuspended in 1 ml incubation solution containing 0.3% (wt/vol) ammonium sulfate, 0.05% (wt/vol) potassium sulfate, 0.01% (wt/vol) potassium chloride, and 0.07% (wt/vol) glycine, pH ∼5.2. The samples were centrifuged again under same conditions. Pellets were resuspended in 200 μl incubation solution containing SYTO9 (0.0068 mM) and propidium iodide (0.04 mM) for 15 min at RT. Samples were analyzed immediately after staining in an ApogeeFlow A-40 flow cytometer (Apogee Flow Systems, Hemel Hempstead, United Kingdom), illuminating with a 488-nm laser.
Flow cytometry analysis.
S. islandicus ΔC1C2 cells were infected at an MOI of 3, and samples were taken regularly from both infected and control cultures; 300 μl of culture was added to 700 μl of 100% ethanol and stored at 4°C. When all the samples were collected, the fixed cells were centrifuged at 3,000 × g for 20 min and resuspended in 1 ml buffer containing 10 mM Tris (pH 7.5) and 10 mM MgCl2. The samples were centrifuged again under same conditions. Pellets were resuspended in 150 μl fresh staining solution containing mithramycin (100 μg/ml) and ethidium bromide (20 μg/ml) for 1 h. At all steps, the samples were kept cold. Samples were analyzed in an ApogeeFlow A-40 flow cytometer (Apogee Flow Systems, Hemel Hempstead, United Kingdom), illuminating with a 405-nm laser.
TEM.
Samples (5 μl) containing SMV1 or Sulfolobus cells infected with SMV1 were adsorbed onto carbon-coated copper grids for 5 min and stained with 2% (wt/vol) uranyl acetate. Images were recorded using a JEM-1010 transmission electron microscope (TEM) (JEOL, Tokyo, Japan) with a Gatan digital camera 792.
RESULTS AND DISCUSSION
Closely related Sulfolobus strains exhibit different susceptibilities to SMV1.
To find a suitable host for studying the life cycle of SMV1, we infected a range of well-established lab strains of Sulfolobus with an MOI of 5 to test for susceptibility. Whereas S. islandicus HVE10 and S. islandicus LAL14/1 showed low or no susceptibility to SMV1 as indicated by similar growth rates of the virus-treated and the control cultures, S. islandicus REY15A and the deletion mutant S. islandicus ΔC1C2 appear to be highly susceptible to SMV1 infection (Fig. 1A and B). REY15A displayed observable growth retardation 14 h postinfection (hpi), whereas ΔC1C2 displayed growth retardation already at 6 hpi. This suggests that strain ΔC1C2 has a higher susceptibility to SMV1 than REY15A. The ΔC1C2 strain was derived from REY15A carrying an ∼160-kbp deletion. The deleted region includes the type I-A clustered regularly interspaced short palindromic repeat (CRISPR) system, the two repeat-spacer arrays, some toxin-antitoxin genes, and hypothetical genes, which likely confers some immunity to SMV1. Interestingly, cultures of S. solfataricus 5E6 show growth retardation between 14 and 23 hpi, at which point the cultures start growing again with a growth rate comparable to that of the control (Fig. 1B). SMV1 has been confirmed by plaque assay to reproduce in 5E6 (data not shown). Investigation of the recovery of 5E6 was not pursued in this work but could prove interesting for future studies investigating whether SMV1 has host-dependent life cycle traits.
FIG 1.
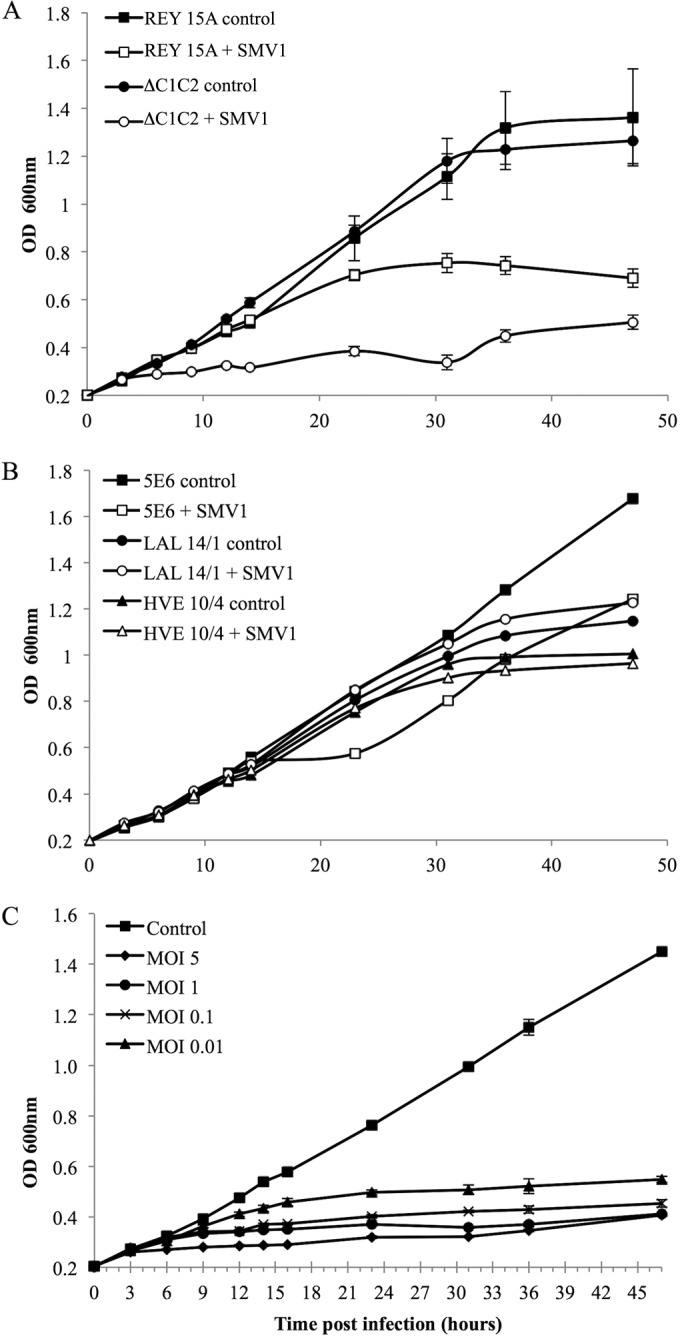
(A and B) Susceptibility of different Sulfolobus strains to SMV1 infection. The OD600 was measured over a time period of 48 h in cultures with (open symbols) or without (filled symbols) the addition of SMV1 (MOI of 5). Cultures were incubated in triplicates at 78°C. (C) Growth inhibition of S. islandicus ΔC1C2 by SMV1 infection at different MOIs (0.01 to 5). Cultures were incubated in triplicates at 78°C.
Based on the results, we selected ΔC1C2 as the host for further investigations into viral life cycle traits of SMV1. First, we infected ΔC1C2 cultures at a range of MOIs (0.01 to 5) to test the effect of MOI on growth retardation. The MOIs were calculated based on viral titers enumerated by plaque assay as PFU/ml. Figure 1C shows that the higher the MOI, the sooner the growth retardation occurs; at an MOI of 5 (when ∼99% of the cells are infected), growth retardation is observed at 6 hpi, compared to 12 hpi at an MOI of 0.01 (when ∼1% of the cells are infected).
SMV1 adsorption is rapid.
To gain insights into the initial stages of SMV1 entry, we followed the kinetics of SMV1 adsorption to ΔC1C2 cells. The adsorption was very efficient, with 50% of virions being bound to cells within 1 min of infection (Fig. 2). Further incubation of the virus in the presence of the host cells resulted in additional virion binding: ∼80% of virions were bound within 20 to 30 min postinfection. All adsorption assays were conducted under optimal growing conditions for the ΔC1C2 host cells, i.e., at 78°C and pH 3.5. To ensure that the observed loss of virus titer was not due to high temperature and/or acidic effects but indeed could be attributed to virus adsorption, we performed a cell-free control experiment in which the same amount of SMV1 as used for the infection was added to ΔC1C2 growth medium. The virus titer of the control did not change over the 30 min of incubation (conditions were as described above). The adsorption rate (calculated as 7 × 10−9 ml min−1 at 1 min postinfection) was very rapid and comparable to the even faster adsorption rate observed for SIRV2 (9). An adsorption assay using a 10-fold-higher MOI resulted in a similar pattern (data not shown), resembling what has been observed previously for SIRV2 (9). SMV1 and SIRV2 are the only two hyperthermophilic archaeal viruses for which the adsorption rate is known. The only other group of archaeal viruses for which adsorption rates have been studied are the viruses of halophilic Archaea, which often bind to their hosts extremely slowly; e.g., 30% adsorption over 3 h is observed for His1 (25, 26). The high adsorption rates of hyperthermophilic viruses are hypothesized to minimize the time they spent in the hostile extracellular environment with boiling temperatures and acidic pH (9).
FIG 2.
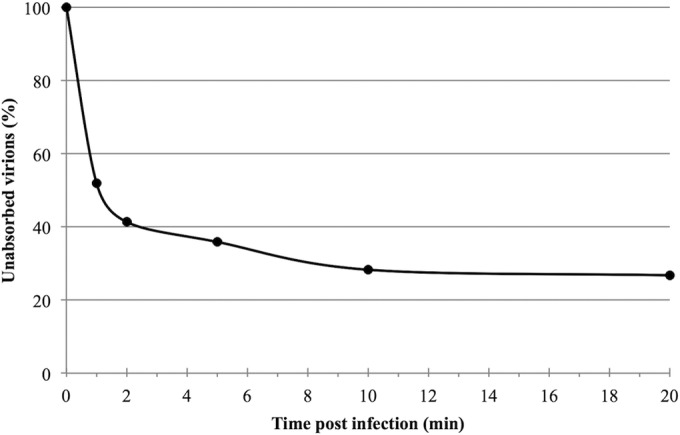
Kinetics of SMV1 adsorption. Cells were infected with SMV1 using an MOI of 0.1 at 78°C. The number of unbound virus particles was determined at different time points postinfection as described in Materials and Methods.
SMV1 exhibits three distinct association modes.
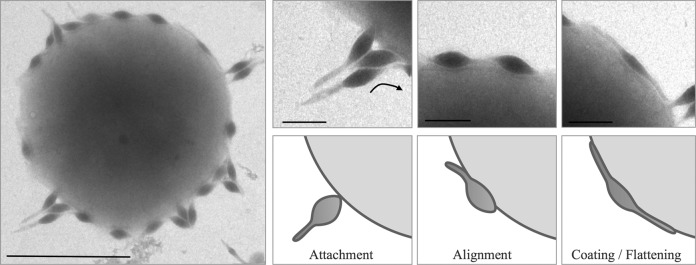
Very little is known about the initial infection stages of archaeal viruses, whereas studies of viruses infecting the other domains of life have revealed three common entry mechanisms: penetration, membrane fusion, and endocytosis (27). The SMV1 virion is amenable to lipophilic dye staining indicative of a lipid component on the surface (K. B. Uldahl and X. Peng, unpublished data). Other spindle-shaped viruses, such as SSV1 and SSV2, have well-established lipid envelopes, providing the potential of fusion entry. To gain insight, we followed the interaction of SMV1 and ΔC1C2 cells by transmission electron microscopy (TEM) just after infection. In general, we observed three distinct modes of viral association with the host cell, i.e., nosecone association, body association, and body/tail association (Fig. 3), possibly occurring sequentially. The first stage appears to be the initial attachment to the host cell; the attachment occurs at the nose-like end, and from the TEM images the receptor appears to be highly abundant, as >30 virions can be attached to a single cell at the same time. The second stage is referred to as the alignment stage. It appears that the virion aligns along the host cell; initially the main body is adsorbed along the surface of the host, while the tail is still unabsorbed. At some point the whole virion, including the tail, is absorbed along the host surface, and the virus particle is coating the cell. At this stage, the virion appears to be flattened against the host surface, and the spindle morphology “disappears” (coating/flattening) (Fig. 3). Taken together, the presence of a lipid component in SMV1 virions and the attachment modes suggest a possible fusion entry mechanism. However, it is clear that further studies are needed to undoubtedly establish the fusion mechanism.
FIG 3.
Electron micrographs of SMV1 interaction with S. islandicus ΔC1C2 cells. Samples were collected at 10 min postinfection and negatively stained for TEM. Left scale bar, 1 μm; right scale bar, 200 nm. The three different virus-host association modes are indicated: attachment, alignment, and coating/flattening.
SMV1 release does not cause cell lysis.
Upon SMV1 infection, clear growth retardation is observed in ΔC1C2 cells; however, it is not clear whether virus release lyses the cells. We first determined the time required for the infecting SMV1 to release viral progeny. A one-step growth curve revealed a dramatic increase of extracellular virus titer at around 8 hpi; this reached the maximum at around 11 hpi (Fig. 4A), which is comparable to the release time for the related virus STSV1 (16) as well as to that for SIRV2 (11). Moreover, this correlates with the observed delayed growth retardation at an MOI of 0.01 (Fig. 1A); at 8 to 12 hpi, new viral progeny from the first replication cycle would have been released and infected the remaining uninfected cells, at which point the growth of the whole culture would be affected.
FIG 4.
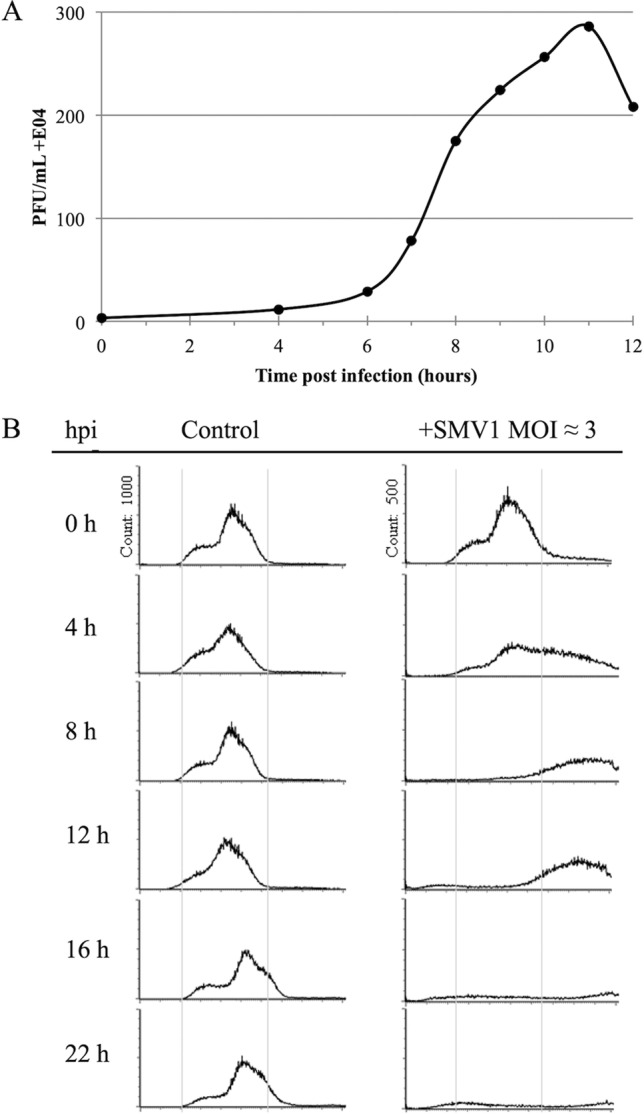
(A) One-step growth curve of SMV1 infection of S. islandicus ΔC1C2. SMV1 was added at an MOI of about 0.1. The PFU are plotted against time (hours). (B) Flow cytometry time course analysis of S. islandicus ΔC1C2 cells infected by SMV1. Left panel, DNA content distribution from an uninfected culture. Right panel, DNA distribution from a culture infected with SMV1 (MOI ≈ 3). The virus was added just before time point 0 h. The experiment was repeated twice.
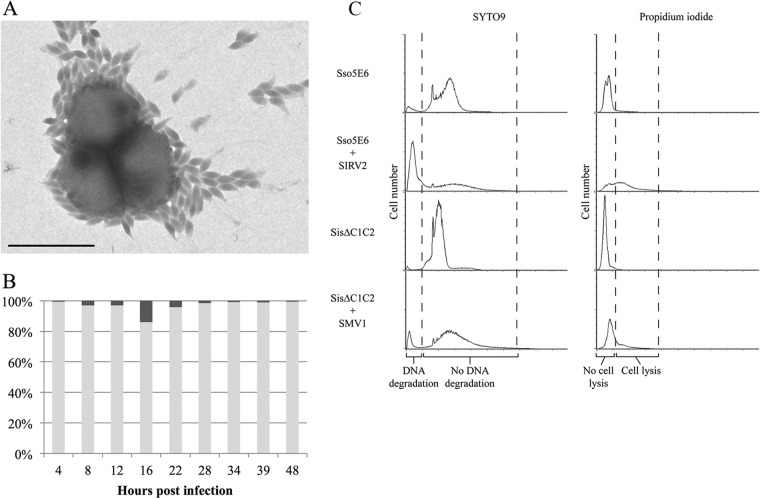
As the lytic virus SIRV2 causes chromosome degradation in the infected host cells, we then investigated the chromosome contents of SMV1-infected cells. The intracellular DNA contents in uninfected and SMV1-infected (MOI of 3) cultures over time were monitored by flow cytometry. The DNA content distributions of the control cultures (Fig. 4B, left) were typical for exponentially growing Sulfolobus cells (28), with a majority of the cells containing 2 chromosomes. In the infected cultures (Fig. 4B, right), cells with a very high DNA content (>2 genomes) started to appear already 4 hpi and continued to increase until 22 hpi, whereas the proportion of cells containing 1 to 2 genome equivalents decreased. At 16 and 22 hpi, a large majority of the cell population had a DNA content above the detection range of the assay (≫ 2 genomes). After extruding from the host cell, SMV1 particles often remain attached to the surface of the cell, and layers of virus particles have occasionally been observed to surround a single or multiple host cells (Fig. 5A). A similar tendency has been observed for STSV1 and host cells (16). This gives rise to particle aggregates with very high DNA contents, which are outside the detection range of the assay of up to 4 genomes. During the 22-h experiment, equivalent to about two viral replication cycles, no significant chromosome degradation occurred in the SMV1-infected cultures. Moreover, we performed a Live/Dead BacLight assay to detect potential membrane disruption of the infected cells. Compared to the uninfected culture, only a negligible increase in the amount of “broken” cells was detected in the SMV1-infected culture over 48 h (Fig. 5B). As a control for the assay, a Sulfolobus culture infected with the lytic virus SIRV2 (10) exhibited a high level of cell lysis (Fig. 5C). Thus, no indications of cell lysis were observed in the SMV1-infected culture, and SMV1 appears to be nonlytic like the related STSV1 (16) and the spindle-shaped fuselloviruses (29, 30).
FIG 5.
(A) Electron micrograph of an S. islandicus ΔC1C2 culture infected with SMV1 (high MOI) at 22 hpi. Scale bar, 1 μm. (B) Time course analysis of the percentage of live (light gray) and dead (dark gray) cells in an S. islandicus ΔC1C2 culture infected with SMV1 (MOI ≈ 3) compared to an uninfected control. (C) Flow cytometry analysis of live/dead staining of S. islandicus ΔC1C2 before and after infection with SMV1 (MOI of 3) at 22 hpi. S. solfataricus 5E6 cells infected with SIRV2 (MOI of 5) were used as a positive control for DNA degradation and cell lysis.
Pleomorphism and stability of SMV1.
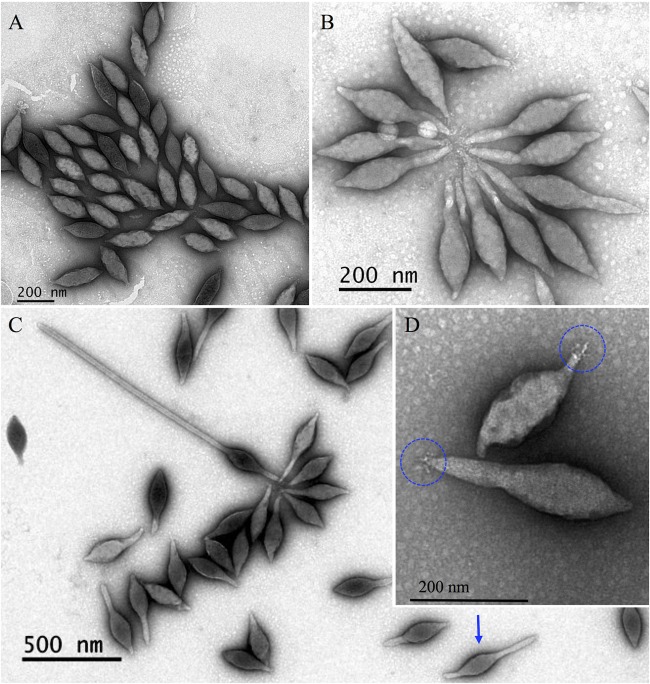
SMV1 particles isolated and purified immediately after release appear spindle shaped with no visible or very short tails (Fig. 6A). Upon release and if left in the cell culture at 78°C, the virions start developing tails; sometimes tail development occurs at both poles, with one tail being shorter than the other. Purified SMV1 particles are often observed in rosette-like structures with virions connected through thin fibers at one pole (Fig. 6B and D). Together this gives rise to a very heterogenic virus population, as seen in Fig. 6C. The closely related ATV displays similar extracellular tail development independent of its host (14). A study measuring ATV virions showed the virion body dimensions of ATV to shift upon tail development: tailless virions exhibited an average maximum width of 119 (±2) nm, while the two-tailed particles showed a maximum width of 85 (±4) nm (14). However, electron micrograph measurements of tailless, one-tailed, and two-tailed SMV1 particles exhibited the same average maximum width of 80 (±4) nm. Thus, a width difference was not observed upon SMV1 tail development. Variation in tail lengths has been reported for both ATV and ATSV, with a maximum reported tail length of ATSV of 720 nm (15). Here, we show that SMV1 can develop tails of more than 900 nm in length (Fig. 6C), more than quadrupling the total virion length. The ability of SMV1 and ATV to undergo major extracellular morphological development is a unique feature of these two archaeal viruses, not observed elsewhere in the virosphere. The reason for the extracellular development of the virions is still unclear.
FIG 6.
Electron micrographs of different forms of SMV1. (A) Tailless SMV1 particles isolated immediately after release. (B and C) SMV1 develops 1 or 2 tails outside the host cell. The development occurs at high temperatures (>75°C). Often one longer and one shorter tail are observed (blue arrow). (D) Only one pole appears to have short tail fibers, which can attach to tail fibers of other virions to form characteristic rosettes (B). All preparations were negatively stained with 2% uranyl acetate.
To gain insights into the mechanism behind the morphological variation of SMV1, we tested the effects of a range of physical and chemical treatments on the morphology and infectivity of SMV1. Highly purified virions were treated with different detergents, different solvents, and proteinase K, as well as a variety of temperature and UV conditions. After each treatment, viral infectivity was determined, and the appearance of the virus particles was assayed by negative-stain TEM. Among the tested conditions, Proteinase K and boiling clearly induced the development of one or two tails at the poles of the virion body, whereas the effect of other treatments was not obvious (see Table S1 in the supplemental material). This suggests that the tail development involves conformational change or reorganization of structural proteins, which can be triggered by environmental conditions such as boiling temperatures.
Surprisingly, SMV1 particles demonstrated extreme stability under a range of the conditions tested above, including freezing without cryoprotectants, which had no effect on infectivity and appearance (see Table S1 in the supplemental material). Moreover, a very high dose (1 J/cm2) of UV irradiation could not completely inactivate SMV1. Plaque assays suggested a 100% inactivation of SMV1 by the high (1-J/cm2) UV irradiation dose and 98% inactivation by a lower UV irradiation dose (40 mJ/cm2, the European standard for the inactivation of most pathogens [31]). However, incubation of 50 ml ΔC1C2 cells with 100 μl of the high-dose UV-treated virus suspension resulted in the production of infectious SMV1 particles at 6 hpi, and a virus titer of 103 PFU/ml was observed by plaque assay at 8 to 10 hpi (data not shown). This indicates that the initially added virus had reproduced. Thus, a complete inactivation had not occurred even at the very high UV dose.
In addition, the UV exposure experiment makes us very cautious when interpreting infectivity by plaque assay. If no infectivity is seen by plaque assay it should not be interpreted as an absolute result but should be followed up by other infectivity assays. Finally, we want to stress the difficulties in estimating precise titers of infectious virus particles. As seen in Fig. 3, more than 30 virus particles can attach to one cell but will give rise to only one plaque. Thus, plaque assays are far from a precise enumeration method, at least for SMV1. Also, we have used a nanoparticle analyzer to estimate the nanoparticle count in a highly purified virus suspension, which gave a 10-fold-higher particle count than that by plaque assay, similar to the particle/PFU ratio estimated with quantitative PCR (qPCR) analysis (K. B. Uldahl and X. Peng, unpublished data). Another study enumerated the spindle-shaped virus SSV9 by plaque assay and qPCR and similarly found a 10-fold difference (32), suggesting this to be a general observation for spindle-shaped viruses.
Concluding remarks.
Our study provides valuable insight into the initial infection stages of one of the large spindle-shaped archaeal viruses, SMV1. Our observations suggest that SMV1 virions adsorb to host cells through three distinct modes, possibly leading to fusion entry. Interestingly, we observed more than 30 virions attached to the surface of a single host cell, indicating a relative abundance of viral receptors. Upon infection, the replication of the virus retarded host growth but did not cause lysis of the host cells. Similar to ATV, SMV1 shows remarkable virion plasticity by developing a tail(s) independent of the host, an extraordinary feature observed only for these two hyperthermophilic spindle-shaped viruses. Moreover, SMV1 particles demonstrate an exceptional resilience to a range of harsh conditions, especially at extremes of temperature and UV irradiance. The work presented here provides a good basis for future studies into the viral traits of the large spindle-shaped archaeal viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Danish Archaea Centre, Copenhagen, for stimulating discussions and helpful advice.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00075-16.
REFERENCES
- 1.Menzel P, Gudbergsdottir SR, Rike AG, Lin L, Zhang Q, Contursi P, Moracci M, Kristjansson JK, Bolduc B, Gavrilov S, Ravin N, Mardanov A, Bonch-Osmolovskaya E, Young M, Krogh A, Peng X. 2015. Comparative metagenomics of eight geographically remote terrestrial hot springs. Microb Ecol 70:411–424. doi: 10.1007/s00248-015-0576-9. [DOI] [PubMed] [Google Scholar]
- 2.Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6:81–93. doi: 10.1038/ismej.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 4.Mojica KD, Brussaard CP. 2014. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol Ecol 89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 5.Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2012. Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol 14:426–440. doi: 10.1111/j.1462-2920.2011.02603.x. [DOI] [PubMed] [Google Scholar]
- 6.Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol Rev 35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 7.Pietila MK, Demina TA, Atanasova NS, Oksanen HM, Bamford DH. 2014. Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol 22:334–344. doi: 10.1016/j.tim.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Prangishvili D, Stedman K, Zillig W. 2001. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol 9:39–43. doi: 10.1016/S0966-842X(00)01910-7. [DOI] [PubMed] [Google Scholar]
- 9.Quemin ER, Lucas S, Daum B, Quax TE, Kuhlbrandt W, Forterre P, Albers SV, Prangishvili D, Krupovic M. 2013. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol 87:13379–13385. doi: 10.1128/JVI.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci U S A 106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okutan E, Deng L, Mirlashari S, Uldahl K, Halim M, Liu C, Garrett RA, She Q, Peng X. 2013. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol 10:875–885. doi: 10.4161/rna.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prangishvili D, Forterre P, Garrett RA. 2006. Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 13.Iverson E, Stedman K. 2012. A genetic study of SSV1, the prototypical fusellovirus. Front Microbiol 3:200. doi: 10.3389/fmicb.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prangishvili D, Vestergaard G, Haring M, Aramayo R, Basta T, Rachel R, Garrett RA. 2006. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol 359:1203–1216. doi: 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 15.Hochstein R, Bollschweiler D, Engelhardt H, Lawrence CM, Young M. 17 June 2015. Large tailed spindle viruses of Archaea: a new way of doing viral business. J Virol doi: 10.1128/JVI.00612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang X, Chen L, Huang X, Luo Y, She Q, Huang L. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erdmann S, Le Moine Bauer S, Garrett RA. 2014. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol 91:900–917. doi: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 18.Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, She Q, Garrett RA, Huang L, Lin L. 2014. A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18:51–60. doi: 10.1007/s00792-013-0591-z. [DOI] [PubMed] [Google Scholar]
- 19.Haring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. 2005. Independent virus development outside a host. Nature 436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- 20.Gudbergsdottir S, Deng L, Chen ZJ, Jensen JVK, Jensen LR, She QX, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendorfer M, Kristjansson JK. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol 16:609–628. [Google Scholar]
- 22.Jaubert C, Danioux C, Oberto J, Cortez D, Bize A, Krupovic M, She Q, Forterre P, Prangishvili D, Sezonov G. 2013. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol 3:130010. doi: 10.1098/rsob.130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Brugger K, Liu C, Shah SA, Zheng H, Zhu Y, Wang S, Lillestol RK, Chen L, Frank J, Prangishvili D, Paulin L, She Q, Huang L, Garrett RA. 2011. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J Bacteriol 193:1672–1680. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams MH. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY. [Google Scholar]
- 25.Kukkaro P, Bamford DH. 2009. Virus-host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep 1:71–77. doi: 10.1111/j.1758-2229.2008.00007.x. [DOI] [PubMed] [Google Scholar]
- 26.Pietila MK, Atanasova NS, Oksanen HM, Bamford DH. 2013. Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environ Microbiol 15:1674–1686. doi: 10.1111/1462-2920.12030. [DOI] [PubMed] [Google Scholar]
- 27.Dimitrov DS. 2004. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol 2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernander R. 2007. The cell cycle of Sulfolobus. Mol Microbiol 66:557–562. doi: 10.1111/j.1365-2958.2007.05917.x. [DOI] [PubMed] [Google Scholar]
- 29.Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci U S A 89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contursi P, Jensen S, Aucelli T, Rossi M, Bartolucci S, She Q. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627. doi: 10.1007/s00792-006-0017-2. [DOI] [PubMed] [Google Scholar]
- 31.Tang WZ, Sillanpaa M. 2015. Virus sensitivity index of UV disinfection. Environ Technol 36:1464–1475. doi: 10.1080/09593330.2014.994040. [DOI] [PubMed] [Google Scholar]
- 32.Bautista MA, Zhang C, Whitaker RJ. 2015. Virus-induced dormancy in the archaeon Sulfolobus islandicus. mBio 6:e02565-14. doi: 10.1128/mBio.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.