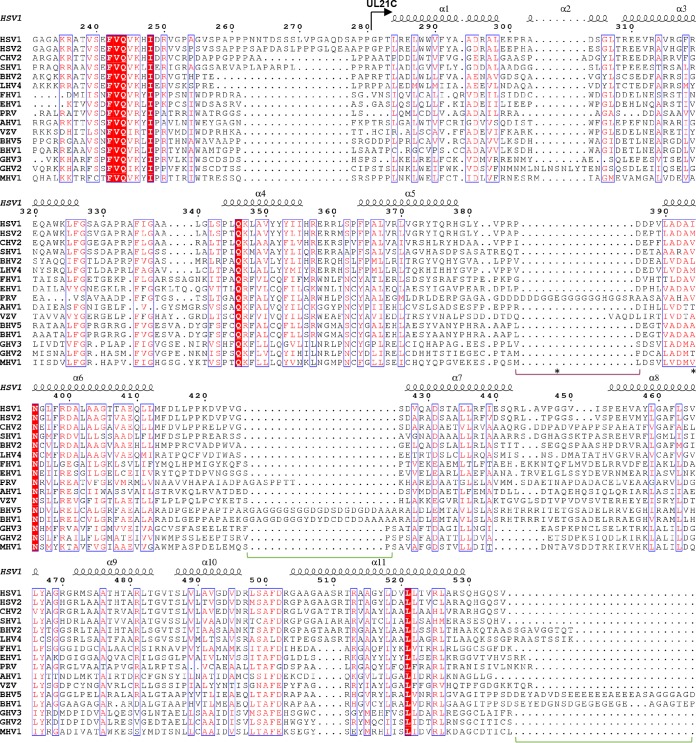
FIG 3.
Sequence conservation in UL21C. Shown is a multiple-sequence alignment of UL21 homologs from 16 alphaherpesviruses. Only the alignment of residues corresponding to residues 231 to 535 of HSV-1 UL21 is shown. The secondary structures of HSV-1 UL21 and the start of UL21C are marked above the alignment. Identical residues are represented by white letters on a red background. Similar residues are represented by red letters boxed in blue. UL21C sequences share 1.2% identity and 8.4% similarity. Insertions in BHV-1, BHV-5, and PRV homologs are indicated by green or purple brackets, respectively, below the alignment. Residues E355 and V375, mutated in PRV Bartha UL21, are indicated by asterisks below the alignment. Alignment was carried out in ClustalW (51) using sequences from the following viruses (with GenBank accession numbers given in parentheses): human HSV-1 (ACM62243), human HSV-2 (AEV91359), cercopithecine herpesvirus 2 (CHV2) (AAU88086), saimiriine herpesvirus 1 (SHV1) (ADO13807), BHV-2 (AAK69349), leporid herpesvirus 4 (LHV4) (AFR32463), feline herpesvirus 1 (FHV1) (ACT88337), equine herpesvirus 1 (EHV1) (AAT67298), PRV (AAA47475), anatid herpesvirus 1 (AHV1) (ABO26208), varicella-zoster virus (VZV) (AAT07796), BHV-5 (AAR86141), BHV-1 (CAA88112), gallid herpesvirus 3 (GHV3) (AEI00223), gallid herpesvirus 2 (GHV2) (AAF66756), and meleagrid herpesvirus 1 (MHV1) (AAG45758). The figure was made in ESPript (52).