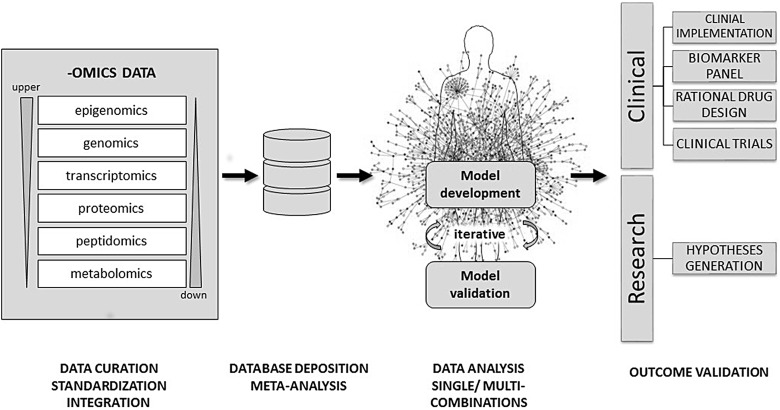
Fig. 1.
Proposed workflow for a data-driven approach in multi-omics data integration. After data acquisition via different high-throughput omic platforms, raw data can be stored locally in the owner's database and be pre-processed (data cleaning, filtering, normalization, reduction, etc.). After the pre-processing steps the data are matched with current reference repositories (data curation). The latter metadata can be then deposited in a different database that only displays statistically relevant features, which is much more amenable for collaborative use for researchers in a common project. Single and/or multiple combinations can be used in order to integrate data coming from up- and downstream levels and then used to develop models that together try to mimic the cell environment and represent their own interactome. The state-of-art model would consider simultaneously any network topology, molecular interaction and statistical relevance in order to provide the most robust representation of the cell dynamics when undergoing disease. Every new model requires confirmation by validating some selected molecular features using in vivo or in vitro experiments (immunohistochemistry, qRT-PCR, ELISA, etc.) This is an iterative step, in which to obtain a final model, it could involve several cycles of incrementing new data and testing its validity until an optimal phase is reached where the model is considered suitable for scientific scrutiny.