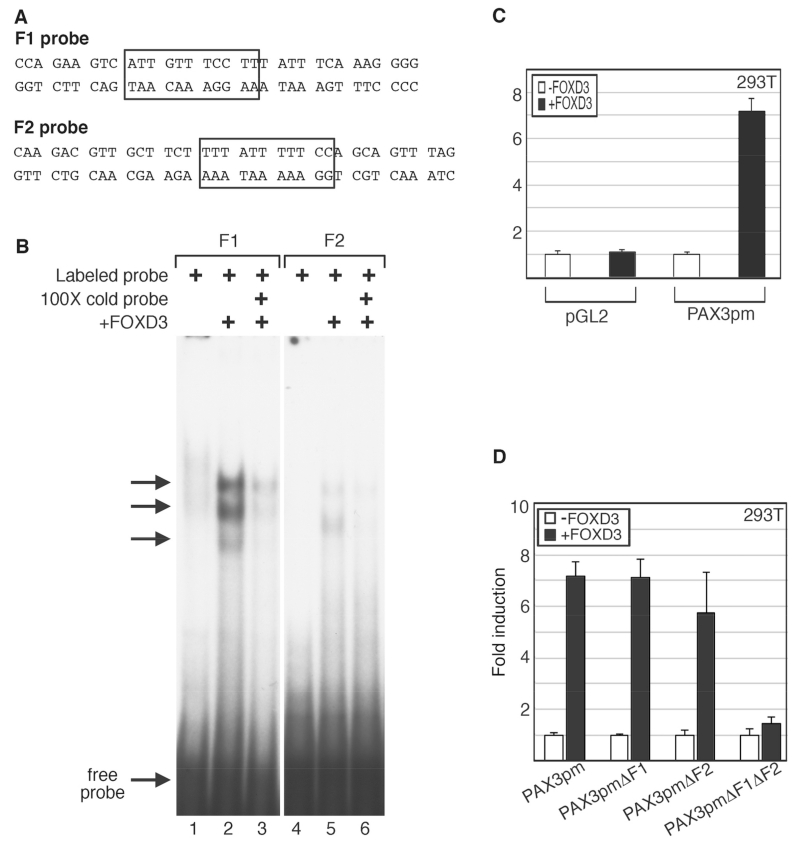
Fig. 5.
FOXD3 binds to the F1 and F2 elements in the PAX3 promoter and can drive expression from these elements in cells. (A) Sequences for the probes used for the EMSA analysis, with the putative FOX binding sites indicated by boxes. (B) EMSA analysis for FOXD3 binding of PAX3 promoter sequences in vitro. Labeled probe containing the PAX3 promoter F1 site (lanes 1-3) or F2 site (lanes 4-6) were mixed with 293T lysate without (lanes 1, 4) or with (lanes 2, 3, 5, 6) exogenous FOXD3 protein. Binding of the labeled probe was inhibited with cold probe competition (lanes 3, 6). (C) FOXD3 promotes expression from the PAX3 promoter. Empty vector (pGL2) or PAX3pm reporter constructs were transfected into 293T cells without (white bars) or with (black bars) FOXD3 expression constructs. (D) FOXD3 drives PAX3 promoter activity in 293T cells through the F1 and F2 sites. Reporter constructs containing the PAX3 genomic region (shown in Fig. 3A and B), driving the expression of luciferase without mutations in putative FOX sites (PAX3pm) or with mutations in F1 (PAX3pmΔF1), F2 (PAX3pmΔF2), or both (PAX3pmΔF1μΔF2), were transfected into 293T cells without (white bars) or with (black bars) a FOXD3 expression construct. For luciferase assays shown in C and D, fold induction was calculated for each sample by the measurement of arbitrary light units produced divided by light units obtained from samples transfected with vector without FOXD3. Each bar is n=9, with standard error of the mean as shown.