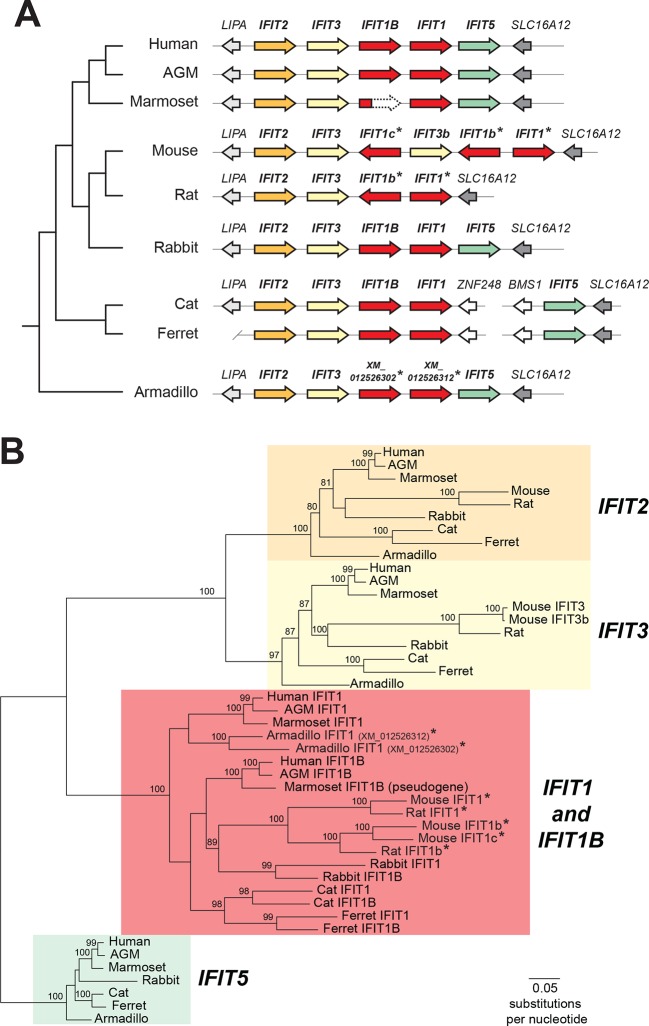
Figure 1. Discordant signatures of synteny and phylogeny for mammalian IFIT genes.
(A) Alignment of the IFIT gene locus from several mammalian genomes. Colored arrows indicate intact IFIT genes and grey arrows indicate neighboring syntenic genes. At left is a schematic phylogenetic tree showing the relatedness of the indicated species. In carnivores such as cats and ferrets, the IFIT locus is split with IFIT5 in one chromosomal location and the remainder of the IFITs in another. In the marmoset genome, the IFIT1B gene has been pseudogenized by frameshift and nonsense mutations, as indicated by the dashed white arrow. Note that the nomenclature for the IFIT genes marked with asterisks is as previously proposed; we suggest a revised nomenclature scheme for those genes in this report from Figure 2 onwards. A version of this figure, with revised gene names and coloring, is found in Figure 2—figure supplement 4. (B) Maximum likelihood phylogenetic tree generated using an alignment of the entire gene sequence of the indicated IFITs. Bootstrap values greater than 80% are shown along the supported branch. Gene names with asterisks next to them have been revised from Figure 2 onward. Accession numbers for all sequences are found in Supplementary file 1.