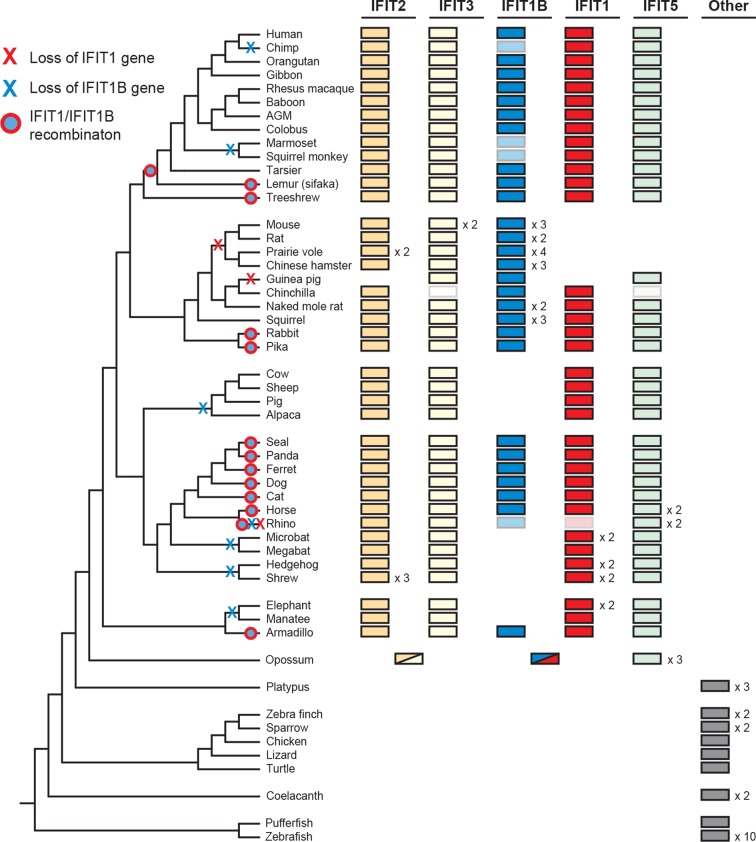
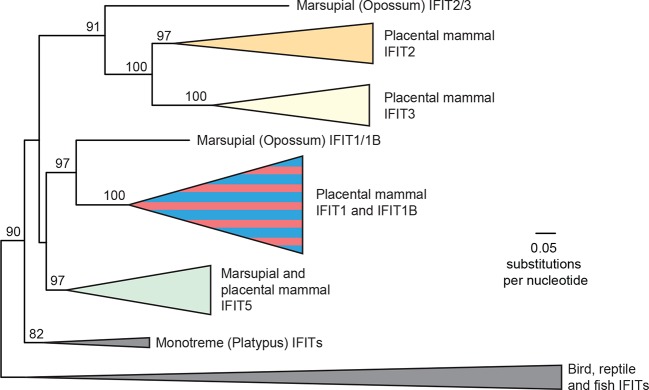
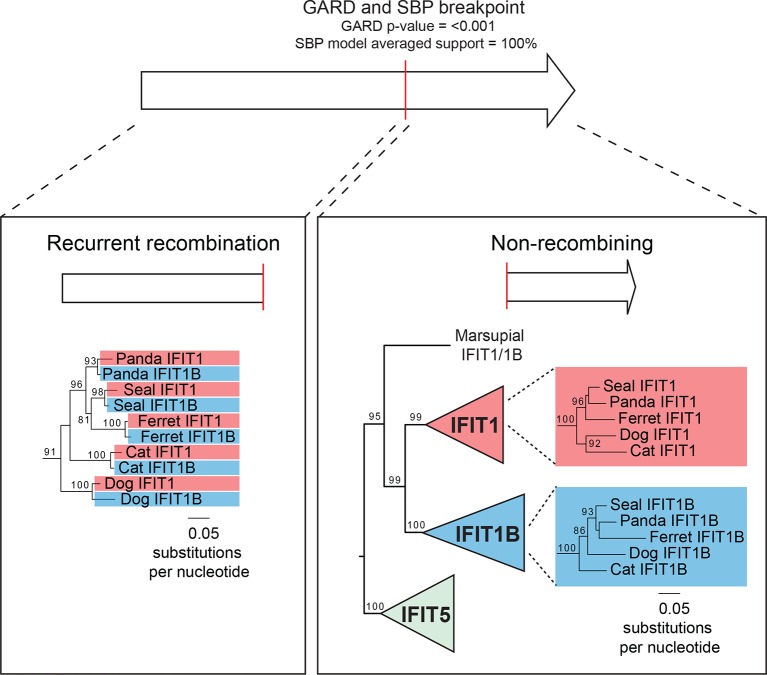
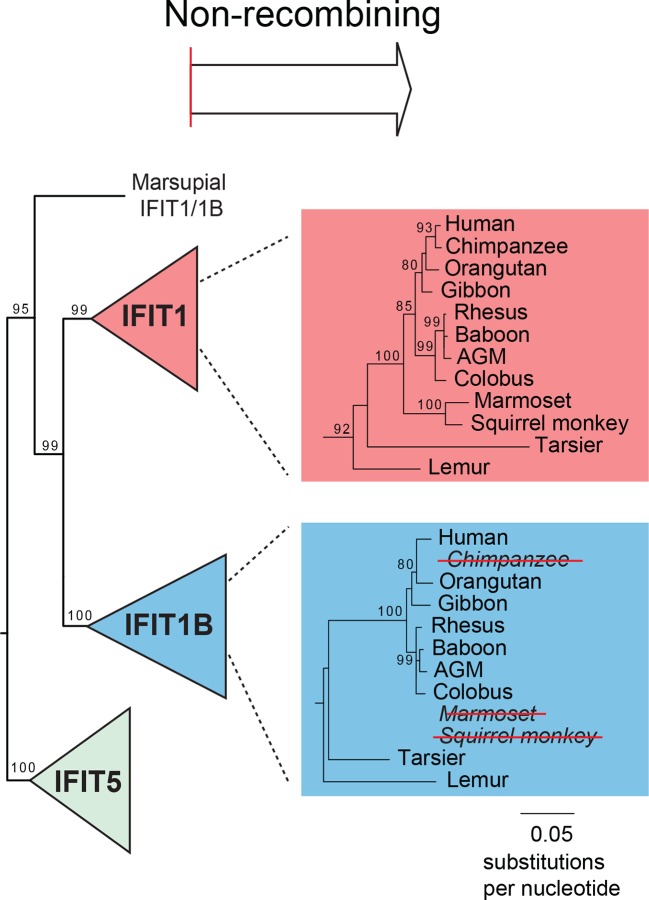
Figure 3. Widespread gene birth, gene loss and gene conversion in vertebrate IFIT genes.
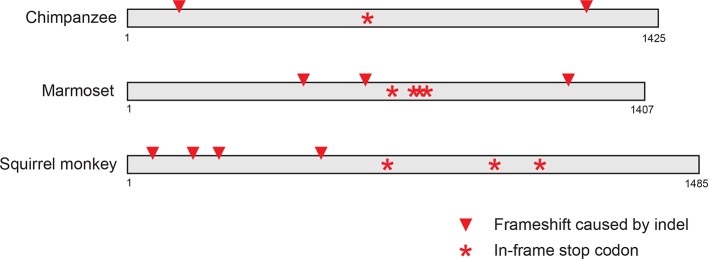
Summary table of the IFIT gene repertoire of 51 vertebrate species (see complete phylogeny in Figure 3—figure supplement 1 as well as expanded regions of trees built from the 5' and 3' end of the IFIT gene alignment in Figure 3—figure supplements 2 and 3). Colored boxes indicate the presence of a given IFIT gene sequence, with multiple copies indicated as a number to the right of the colored box. Lightened boxes indicate that only a partial or pseudogenized copy of the gene can be found in the genome. On the phylogenetic tree to the left, X's indicate deletion or pseudogenization of either IFIT1 or IFIT1B genes from that lineage (see Figure 3—figure supplement 4 for primate examples). Also shown are predicted occurrences of gene conversion (red-around-blue circles) between IFIT1 and IFIT1B sequences based on discordance between phylogenetic trees generated from 3' or 5' regions of the genes (for carnivore examples see Figure 3—figure supplement 2). Accession numbers for all sequences are found in Supplementary file 1.