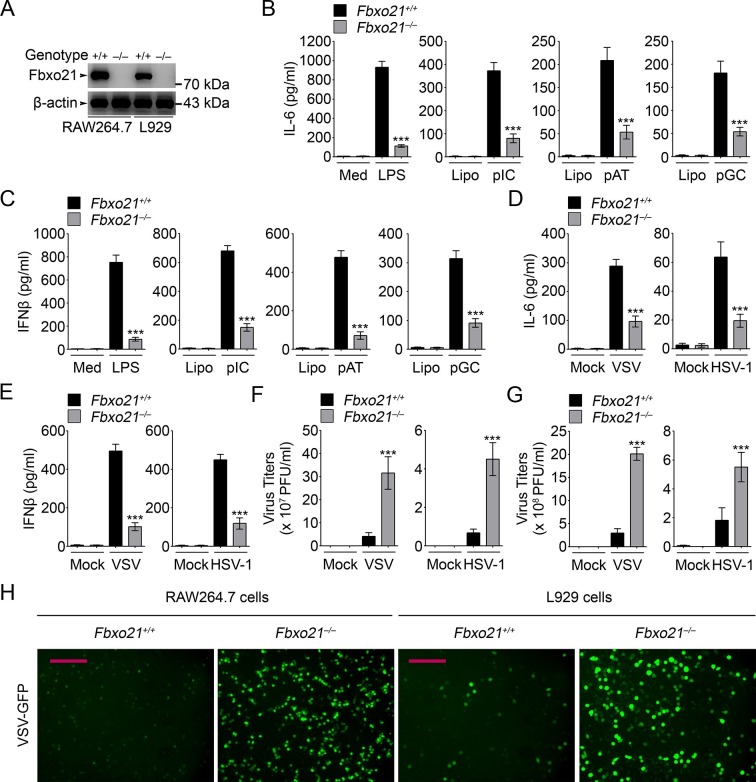
Figure 2. Deficiency of Fbxo21 impairs antiviral innate response.
(A) Confirmation of Fbxo21 depletion. Fbxo21 expression in single-clone-derived RAW264.7 cells and L929 cells established by CRISPR-Cas9 editing was examined by immunoblot. One representative experiment of three was shown. (B, C) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (1 × 105 cells per 24-well) were treated with or without 100 ng/ml LPS (for 6 hr), or transfected with 2.5 μg/ml of liposome-packaged poly (I:C), poly (dA:dT) or poly (dG:dC) (for 8 hr). IL-6 (B) and IFNβ (C) concentrations in supernatant were determined by ELISA. Med, medium; CpG, CpG-ODN; pIC, poly (I:C). (D–G) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (D–F) or L929 cells (G) were infected with or without (Mock) VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated for 8 hr (D, E) or 12 hr (F, G). IL-6 (D) and IFNβ (E) concentrations in supernatant were determined by ELISA. The titers of viruses in cells were determined by plaque formation assay (F, G). In (B–G), error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p < 0.001; versus corresponding Fbxo21+/+ group). (H) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells were infected with or without (Mock) VSV-GFP virus (MOI = 1) for 8 hr. Then cells were viewed under immunofluorescence microscope. One representative experiment of three was shown. Scale bars: 200 μm.