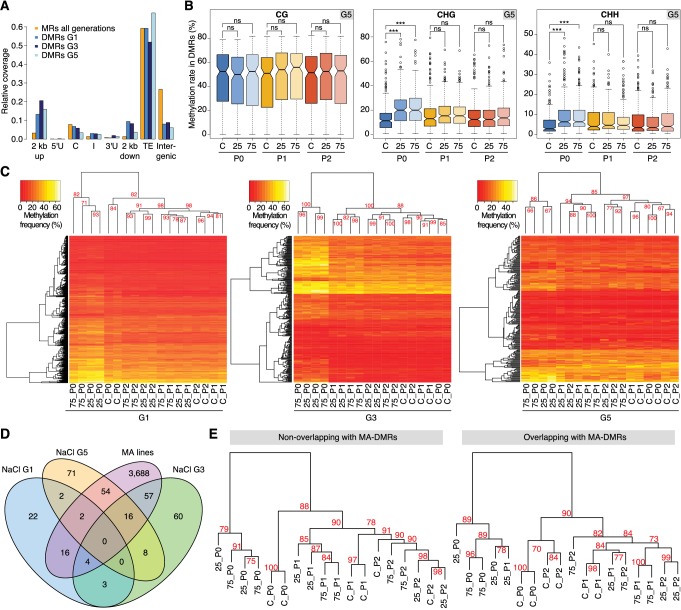
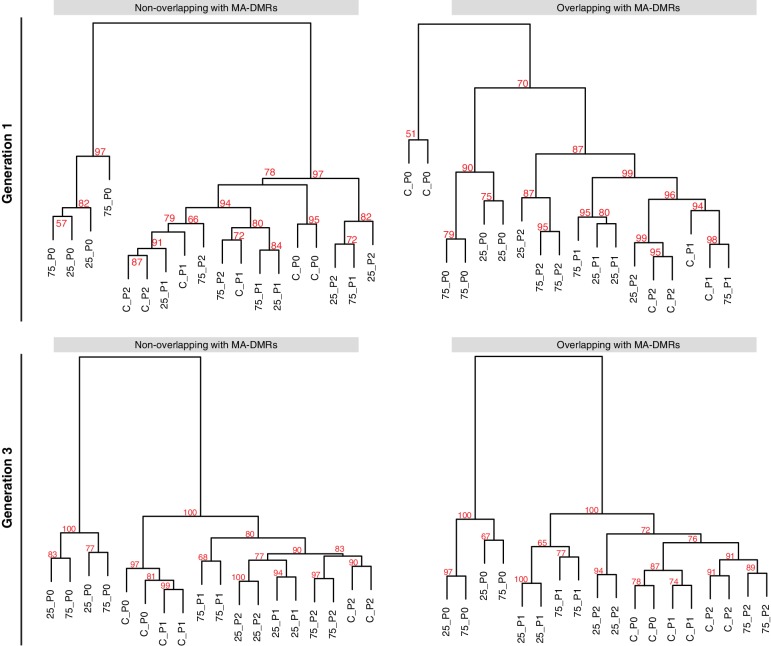
Figure 2. Hyperosmotic stress-induced differentially methylated regions (DMRs) in the absence of stress stimulus.
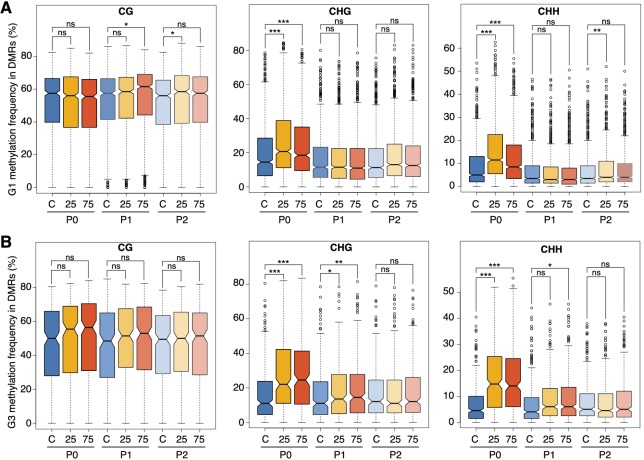
(A) Annotation of cytosines in MRs and DMRs between P0 control and P0 hyperosmotic treated samples in different generations (see Figure 1A). (B) Methylation frequencies by sequence context in DMRs identified between control (P0 of G5), stress-treated (P0 of G5), and the derived P1 and P2 plants (unpaired two-tailed Student’s t-test; ***p<0.001, ns p>0.05). Horizontal bar corresponds to median, whiskers indicate entire 95th percentile. (C) Complete linkage clustering of samples from different generations based on DMR methylation frequencies. Methylation frequency of cytosines contained in each DMR were averaged, and only DMRs covered in all samples were considered. Numbers in red indicate approximately unbiased (AU) p-values (x100), calculated with pvclust. (D) Overlap (including 500 bp flanking windows) of DMRs between P0 control and stress-treated samples from G1, G3 and G5. Overlap with DMRs from a previous analysis of mutation accumulation (MA) lines (Hagmann et al., 2015) is also shown. (E) Clustering of DMRs between P0 control and stress-treated samples in G5 according to overlap with MA-DMRs. C, control, 25, 25 mM NaCl and 75, 75 mM NaCl.