Fig. 4. IFNG-inducible ERVs are pervasive in mammalian genomes.
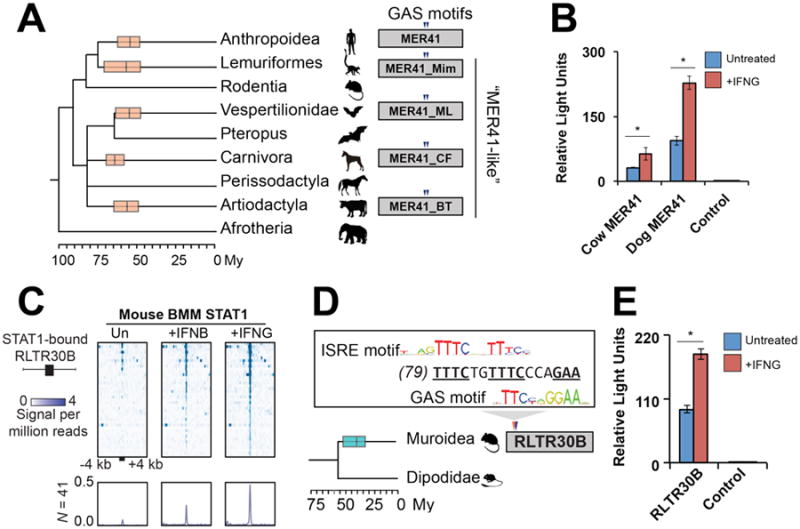
A) A consensus mammalian species phylogeny overlain with boxplots (median and 25th/75th percentiles) depicting the estimated age of MER41-like amplifications (18). Triangles depict conserved GAS motifs. B) Luciferase reporter assays of MER41-like LTR consensus sequences from cow and dog (18). C) Heatmap of ChIP-Seq signals centered across STAT1 peak summits within mouse RLTR30B elements. BMM: bone marrow-derived macrophages. Bottom metaprofiles represent average normalized ChIP signal across bound elements. D) Rodent phylogeny overlain with a boxplot depicting the amplification of RLTR30B, as in (A). ISRE: Interferon Stimulated Response Element motif (TTTCNNTTTC) predicted to bind STAT1 in response to IFNB (13). E) Luciferase reporter assay of RLTR30B consensus sequence, as in (B). Time-calibrated phylogenies in (A) and (D) are from (34). * p < 0.05, Student's t-test.
