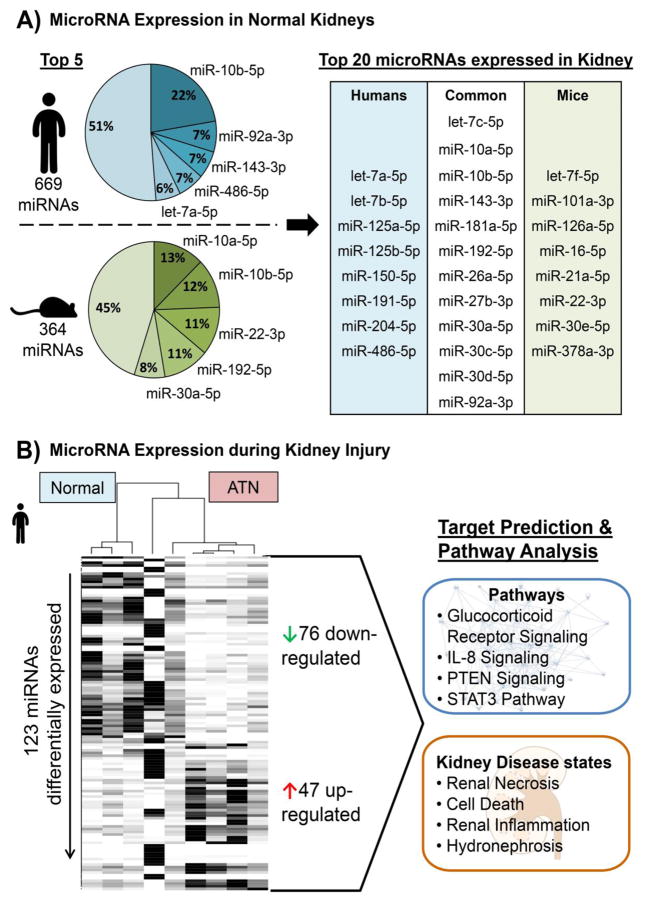
Figure 2. MicroRNA Expression in Healthy and Injured Kidneys.
A) To measure the baseline expression levels of microRNAs in the kidney total RNA was extracted from normal human (n=5) and mice (n=3) kidney samples followed by a small RNA sequencing using the Illumina platform. The normalized mean read counts were ranked by defining read counts >10 as being expressed. The proportions of the top expressed microRNAs were compared within each species as well as between human and mouse and revealed a big overlap of highly abundant microRNAs in the kidneys. B) Total RNA was also extracted and small RNAs were sequenced from kidney samples of patients with acute tubular necrosis (ATN; n=5). Differentially expressed microRNAs between normal and ATN kidney were identified using the DEseq2 algorithm including an adjustment for age and gender of the patients. This resulted in 47 up- and 76 down-regulated microRNAs in ATN vs. normal (FDR adjusted p<0.05 and fold-change cut-off +/− 2). Subjecting the 123 total deregulated microRNAs to a target prediction and pathway analysis utilizing Ingenuity Pathway Analysis demonstrated that these miRNAs are associated with specific pathways as well as kidney disease states.