Fig. 2.
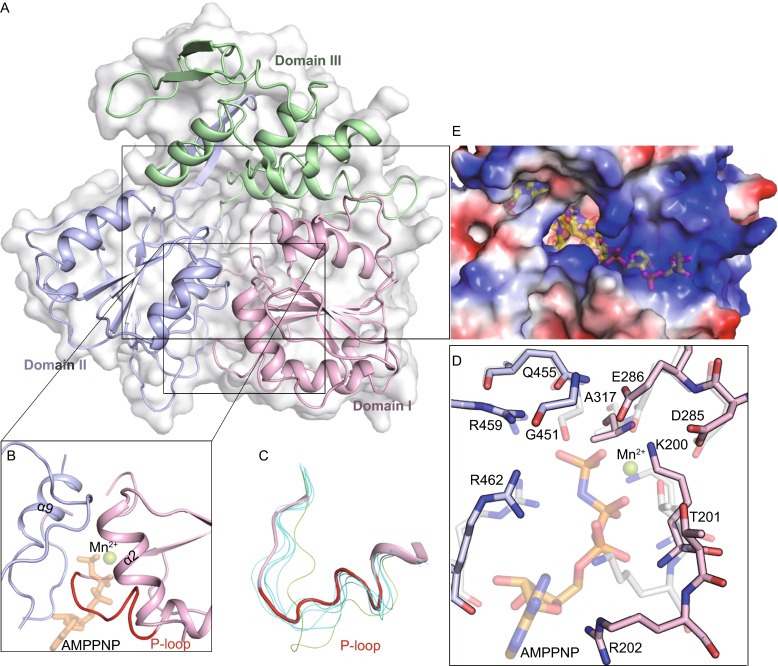
Structural insight into ZIKV helicase. (A) Cartoon and surface representation of the overall fold with the three domains of ZIKV helicase, colored and labeled respectively; (B) The electrostatic surface representation showing the tunnel for potential RNA binding. Positive potentials are colored blue and the negative are colored red. The putative position of the nucleic acid is marked as semi-transparent sticks. The model was obtained by superposition with the DENV-4 helicase in complex with ssRNA (PDB code 2JLV). (C) A clear view of the NTPase active site. The positions of putative nucleotide substrate (as sticks) and Mn2+ (as sphere) are marked semi-transparently by superposition with the DENV-4 helicase bound to AMPPNP and Mn2+ (PDB code 2JLR). P-loop is shown in red. (D) Isolated P-loops are shown by superimposing the structures of 7 flavivirus apo helicases. ZIKV helicase is in red ribbon and the others are shown in finer lines. The P-loop of DENV-4 helicase is colored green. The following structures of helicases with PDB ID in parentheses are included: DENV-2 (2BMF), DENV-4 (2JLQ), JEV (2Z83), KUNV (2QEQ), YFV (1YKS), MVEV (2V8O). (E) Interactions at NTPase active site by superposition of ZIKV helicase (solid) with DENV-4 helicase in complex with AMPPNP and Mn2+ (semitransparent, PDB code 2JLR). Conserved residues are shown as sticks and labeled
