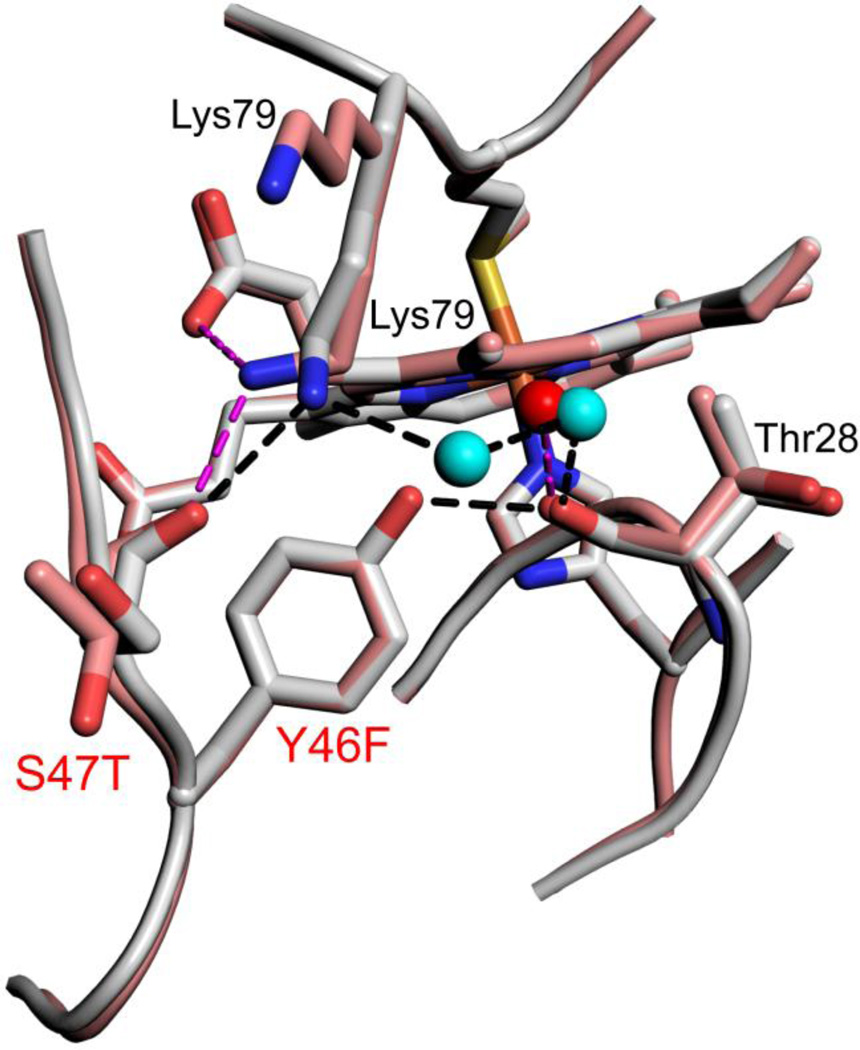
Fig. 5.
Close-up of changes in the hydrogen bond network around position 46 for Hu Cytc (Tyr 46, gray, chain A, pdb code: 3ZCF, [41]) and SM Cytc (Phe46, salmon, chain A, pdb code: 5DFS). Sites of sequence substitutions are labeled red. Other residues involved in the hydrogen bond network have black labels. Both positions of Lys79 in SM Cytc are labeled. Waters in the hydrogen bond network are shown as cyan (Hu Cytc) and red (SM Cytc) spheres. Hydrogen bonds (black dashed lines) for Hu Cytc have the following lengths: Lys79 εN to Ser47 carbonyl-O, 2.84 Å; Lys79 εN to water 2100, 2.76 Å; W2100 to W2043, 2.63 Å; W2043 to Thr28 carbonyl-O, 2.74 Å; Tyr46-OH to Thr28 carbonyl-O, 2.63 Å. Hydrogen bonds (magenta dashed lines) for SM Cytc have the following lengths: Lys79 εN to Thr47 carbonyl-O, 2.76 Å; Lys79 εN to heme propionate D, 3.05 Å; water 362 to Thr28 carbonyl-O, 2.79 Å.