Abstract
The growth arrest-specific 2 (GAS2) was cloned and found to be upregulated in the feces of recurrent CRC patients. This overexpressed GAS2 induced different patterns of gene expressions in CRC cells. Briefly, one cell proliferation marker, Ki-67 antigen (Ki-67), was upregulated in the cells with overexpressed GAS2, “Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer” [1]. Whereas, the expression of another cell proliferation marker, proliferating cell nuclear antigen (PCNA), changed insignificantly [1]. In addition, the mRNA level of one cyclin involving in both cell cycle G1/S and G2/M transitions was also not affected by GAS2 overexpression, “Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A” [2]. The experimental design and procedures in this article can be helpful for understanding the molecular significance of GAS2 in SW480 and SW620 CRC cells.
Specifications Table
| Subject area | Biology |
| More specific subject area | Molecular medicine |
| Type of data | Table, figure |
| How data was acquired | The commercial kit, real-time PCR, SPSS 22.0 for Window software |
| Data format | Filtered, analyzed |
| Experimental factors | CRC patients with different clinical status (recurrence and nonrecurrence) were enrolled. CRC cell lines, SW480 and SW620, were collected for further transfection. |
| Experimental features | Fecal total RNA was purified from these CRC patients with different clinical status and microarray analyses were performed. In addition, PCNA, Ki-67, and CCNA2 were quantified by real-time PCR from SW480 and SW620 CRC cells according to their GAS2 expressions. |
| Data source location | New Taipei, Taiwan, Republic of China |
| Data accessibility | Data are presented in this article |
Value of the data
-
•
Expression levels of certain genes in feces were different between CRC patients without and with recurrence.
-
•
The data would be valuable to correlate the cell proliferation markers and the expression levels of GAS2.
-
•
The data might support the irrelevance of GAS2 in G2 phase of cell cycle.
1. Data
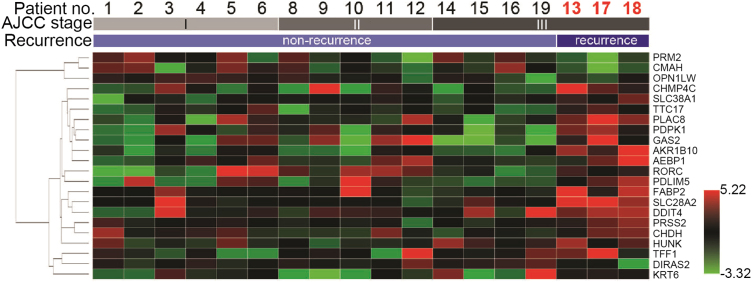
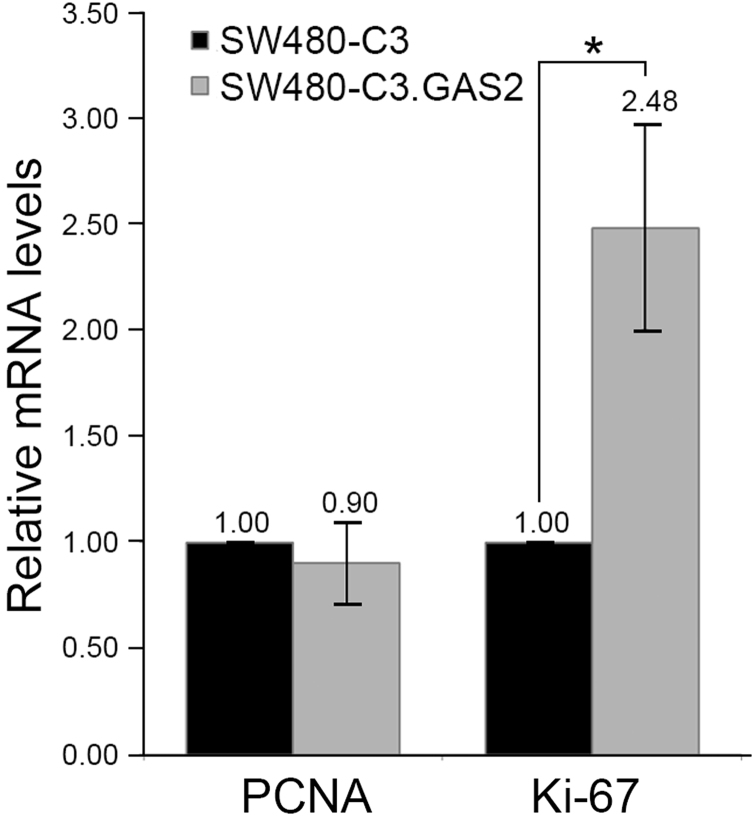
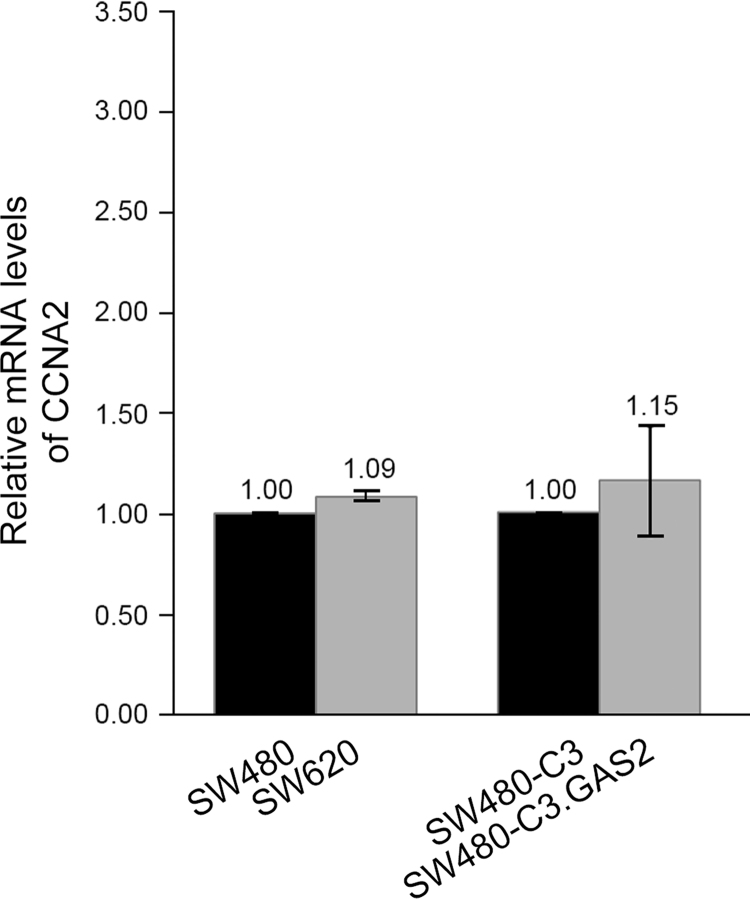
The data shared in this article are the experimental analyses of GAS2 expression in clinical samples, patient׳s feces, and CRC cell lines. The level of GAS2 would be up-regulated in the cases with recurrent CRC by microarray analysis (Fig. 1) [3]. The relative mRNA levels of interested genes (proliferating cell nuclear antigen: PCNA; Ki-67 antigen: Ki-67; cyclin A2: CCNA2) in different cells are presented in Figs. 2 and 3.
Fig. 1.
Clusters of 22 significant genes for recurrent CRC. The relative fold and statistic significance in these assays were set at >1.5-fold and P<0.05 on the similarity between their expressions in cases. Values of high (red) and low (green) expressions were indicated at lower right corner.
Fig. 2.
mRNA levels of proliferation markers in GAS2-overexpressed SW480 cells. Relative value of mRNA levels was indicated at bar top. SW480-C3, SW480 cells with pEGFP C3; SW480-C3.GAS2, SW480 cells with GAS2-contained pEGFP C3. PCNA, proliferating cell nuclear antigen; Ki-67, Ki-67 antigen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAS2, growth arrest-specific-2. SW480, ATCC CCL-228. *P<0.05.
2. Experimental design, materials and methods
2.1. Expression difference in feces of nonrecurrent and recurrent CRC patients
Candidates in feces of recurrent patients were clustered from each comparison in relative to those fecal expressions of non-recurrent patients. The case numbers and stages in these bioinformatic analyses were 15 non-recurrent patients (6 stage I, 5 stage II, and 4 stage III) and 3 recurrent patients (stage III). Candidates in CRC tissues were extracted from GEO: GSE17536, GEO: GSE17537, GEO: GSE17538_GPL570 (Moffitt Cancer Center and Vanderbilt University Medical Center, respectively), GEO: GSE12032 (individual comparison at each stage for non-recurrent vs. recurrent), and GEO: GSE27854 at the public website of Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) [4], [5], [6], [7]. Genes with potentially significant changes were selected by comparing the data from non-recurrent and recurrent CRC tissues. 22 genes were selected due to the cross-compared results from fecal and CRC tissue microarrays.
2.2. The significance of GAS2 in cell proliferation
We established two SW480 cell lines to overexpress GAS2. Firstly, GAS2 cDNA was amplified from a human placenta library (Sigma-Aldrich, St. Louis, MO, USA) (Table 1) and the polymerase chain reaction (PCR) bands for GAS2 were sequenced to confirm gene identity. Then, SW480 cells expressing GAS2 were generated either by lentiviral transduction using the all-in-one tetracycline-inducible plasmid (pAS4.1w.Ppuro-aOn) (Academia Sinica, Taipei, Taiwan) or by stable transfection using a plasmid encoding green fluorescent protein (pEGFP C3) (Takara Bio, Shiga, Japan).
Table 1.
Primers for cloning growth arrest-specific 2 into pAS4.1w.Ppuro-aOn plasmid.
| Official symbol | Accession | Sequence (from 5׳ to 3׳)a | Product size |
|---|---|---|---|
| GAS2 | NM_005256 | F: GCGATCGCGCTAGCATGTGCACTGCTCTGAGCCC | 964bp |
| R: GTTTAAACTCACTTAATTTCCTTCTTAGCCT |
Abbreviations: GAS2, growth arrest-specific 2; F, forward primer; R, reverse primer; pAS4.1w.Ppuro-aOn, all-in-one tetracycline-inducible plasmid.
Underline indicated the AsiSI restriction site at forward primer and the PmeI restriction site at reverse primer; start codon (ATG) and stop codon (TGA; reverse sequence: TCA) were in bold letters.
The relative mRNA levels of PCNA (NM_002592) and Ki-67 (NM_002417) in cells were quantified by quantitative real-time PCR and relative to the mRNA level of GAPDH (NM_002046). As shown in Fig. 2, the level of Ki-67 in SW480-C3.GAS2 was significantly higher than that in control cells without GAS2 overexpression (SW480-C3). Primer sequences used in these gene quantifications were described as following: PCNA (universal probe, #69), 5′-TGGAGAACTTGGAAATGGAAA-3′ for forward primer and 5′-GAACTGGTTCATTCATCTCTATGG-3′ for reverse primer; Ki-67 (universal probe, #73), 5′-CAAGAGGTGTGCAGAAAATCC-3′ for forward primer and 5’-TCACTGTCCCTATGACTTCTGG-3′ for reverse primer. The expression level of GAPDH (universal probe, #60; forward primer, 5′-CTCTGCTCCTCCTGTTCGAC-3′; reverse primer, 5′-ACGACCAAATCCGTTGACTC-3′) was used as an internal control for calibration.
2.3. The correlation of GAS2 in the expression of CCNA2
The molecular marker for G2 phase of cell cycle, CCNA2 (NM_001237), was specific quantified by quantitative real-time PCR. The relative mRNA levels of CCNA2 were also calibrated with that of GAPDH as described before. However, no significant difference was found in the cells with overexpressed GAS2 (Fig. 3). Sequences of primer pair for CCNA2 were 5′-CCATACCTCAAGTATTTGCCATC-3′ (forward primer) and 5′-TCCAGTCTTTCGTATTAATGATTCAG-3′ (reverse primer). The number of universal was #67.
Fig. 3.
mRNA levels of CCNA2 in relation to GAS2 overexpression. Relative value of mRNA levels was indicated at bar top. SW480-C3, SW480 cells with pEGFP C3; SW480-C3.GAS2, SW480 cells with GAS2-contained pEGFP C3. CCNA2, cyclin A2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAS2, growth arrest-specific-2. SW480, ATCC CCL-228; SW620, ATCC CCL-227.
Acknowledgments
We thank Mr. Ting-Liang Weng and Ms. Hsiao-Ting Tseng for their assistance in bioinformatic analysis and gene quantitations. This work was supported by the Grant (Grant no. 103CGH-TMU-05 to C.J. Huang and C.C. Chang) from the Cathay General Hospital and Taipei Medical University.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.05.010.
Appendix A. Supplementary material
Supplementary material
References
- 1.Guzinska-Ustymowicz K., Pryczynicz A., Kemona A., Czyzewska J. Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009;29:3049–3052. [PubMed] [Google Scholar]
- 2.Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Chien C.C., Chang C.C., Yang S.H., Chen S.H., Huang C.J. A homologue of the Drosophila headcase protein is a novel tumor marker for early-stage colorectal cancer. Oncol. Rep. 2006;15:919–926. [PubMed] [Google Scholar]
- 4.Kuo T.Y., Hsi E., Yang I.P., Tsai P.C., Wang J.Y., Juo S.H. Computational analysis of mRNA expression profiles identifies microRNA-29a/c as predictor of colorectal cancer early recurrence. PLoS One. 2012;7:e31587. doi: 10.1371/journal.pone.0031587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoud O., Harrison A., Perperoglou A., Gul A., Khan Z., Metodiev M.V., Lausen B. A feature selection method for classification within functional genomics experiments based on the proportional overlapping score. BMC Bioinf. 2014;15:274. doi: 10.1186/1471-2105-15-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berdiel-Acer M., Berenguer A., Sanz-Pamplona R., Cuadras D., Sanjuan X., Paules M.J., Santos C., Salazar R., Moreno V., Capella G., Villanueva A., Mollevi D.G. A 5-gene classifier from the carcinoma-associated fibroblast transcriptomic profile and clinical outcome in colorectal cancer. Oncotarget. 2014;5:6437–6452. doi: 10.18632/oncotarget.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An N., Yang X., Zhang Y., Shi X., Yu X., Cheng S., Zhang K., Wang G. Cell cycle related genes up-regulated in human colorectal development predict the overall survival of late-stage colorectal cancer patients. Mol. Biosyst. 2016;12:541–552. doi: 10.1039/c5mb00761e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material