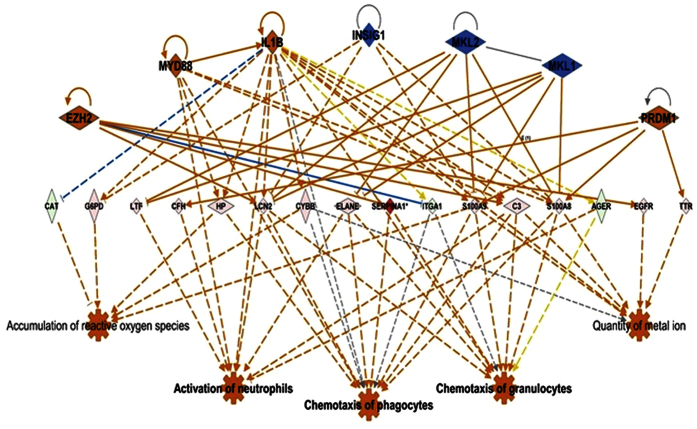
Figure 7. Predicted regulatory network effects identified from differential protein expression profile in lungs of C57BL/6 mice in response S. pneumoniae TIGR4 12 h p.i.
Based on significant up- (shown in pink and red) down- (shown in green) regulation of lung proteins (represented by diamond shape), Ingenuity pathways analysis (IPA) predicted activation of upstream regulators such as EZH2, MYD88, IL-1β and PRDMI (shown in orange) and inhibition of INSIG1, MKL1 and 2 (shown in blue). IPA’s regulator effects algorithm connected the upstream regulators, proteins in our dataset to downstream functions to generate regulator effects hypotheses with a consistency score. The predicted top regulatory network (13.8 consistency score) in response to TIGR4 will impact chemotaxis of granulocytes, phagocytes and activation of neutrophils consistent with the establishment of infection, a delayed response when compared to ∆potABCD 4 h (Fig. 6).