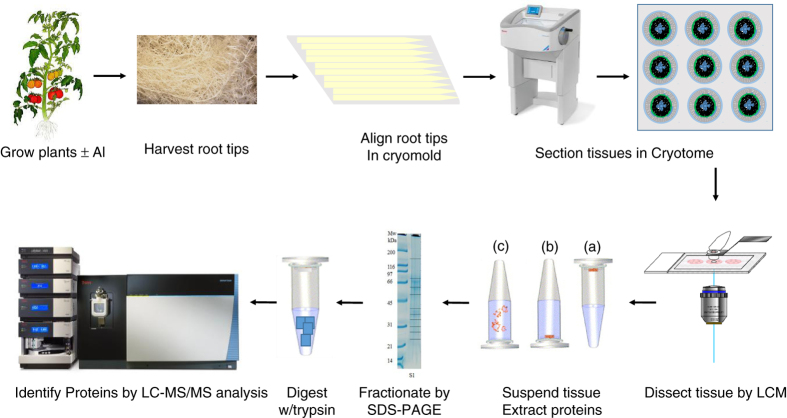
Figure 1.
Workflow of laser capture microdissection single-cell-type proteomics of tomato roots. During step 1, roots were harvested, fixed and embedded in optimal cutting temperature compound (OCT embedding compound). Tissue sections (10 μm in thickness) were cut using a cryostat microtome at −20 °C. During step 2, using a PALM MicroBeam Laser Capture Microdissection (LCM) system, single-cell layer tissues were cut, lifted from the slides and collected into a capture tube using an ultraviolet laser. During step 3, proteins were extracted from the LCM tissues and separated on SDS–PAGE gel (1D), followed by in-gel trypsin digestion (note that the dark lines in the right lane of the electropherogram represent fraction boundaries and not distinct proteins). During step 4, the tryptic peptides were analyzed using nano liquid chromatography-tandem mass spectrometry (LC-MS/MS) and proteins were identified by comparing these spectra against theoretical spectra generated in silico from the unigenes in ITAG2.4 tomato protein database. 1D, one-dimensional; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.