Figure 1.
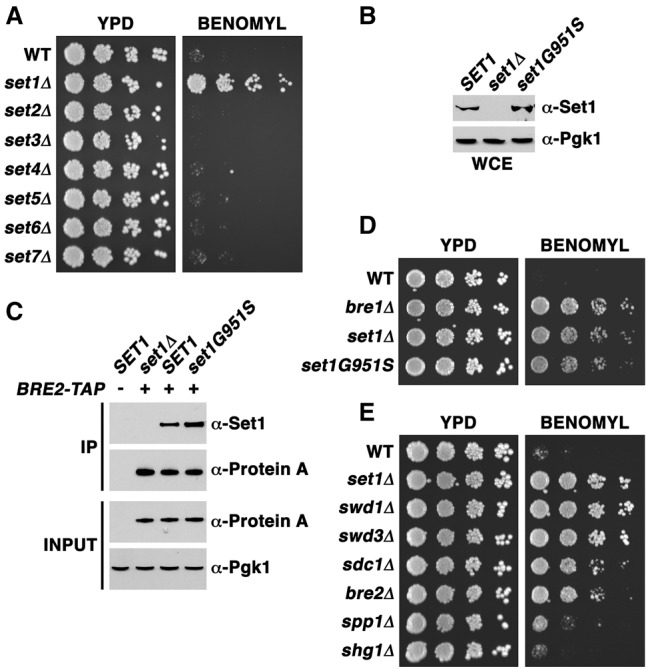
Loss of COMPASS-mediated lysine methylation results in benomyl resistance. (A) A serial fivefold dilution assay of yeast strains with the indicated genotypes was performed to visualize growth phenotypes. Cells were placed onto YPD plates (YPD) with or without 30 µg/mL benomyl, and these plates were placed for 2 d at 30°C. (B) Whole-cell extracts (WCEs) were isolated from strains with the indicated genotypes. The amount of total Set1 protein was assessed by immunoblot analysis using an antibody specific for Set1 (1:100; Santa Cruz Biotechnology, SC-101858). Pgk1 protein levels were used as a loading control. (C) Both wild-type Set1 and Set1G951S proteins similarly associate with the COMPASS component Bre2. Immunoprecipitation (IP) assays were used to pull down Bre2, and the associated levels of Set1 were assessed in the wild-type (WT) and set1G951S strains. Bre2-TAP (α-protein A) and Pgk1 (α-Pgk1) were used as loading controls. (D) bre1Δ, set1Δ, and the catalytically inactive mutant set1G951S were subjected to the same assay as in A. (E) COMPASS mutants were assessed for growth phenotypes using the same serial fivefold dilution assay as in A and D.
