Figure 3.
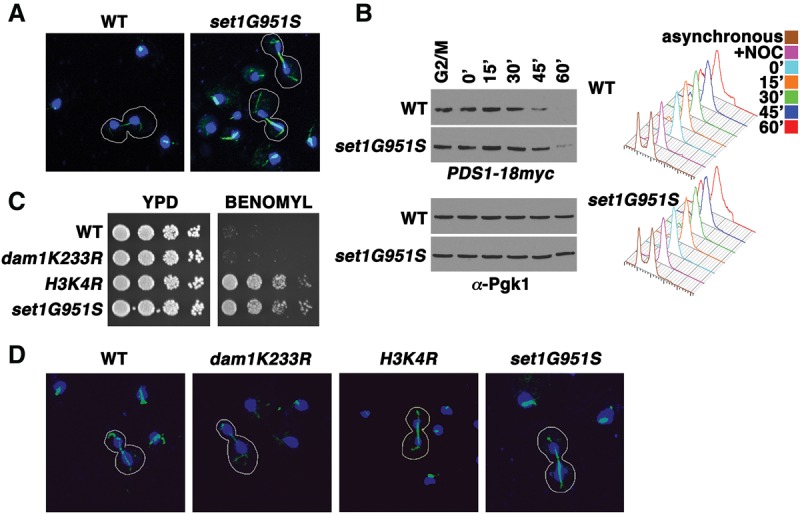
Loss of H3K4 methylation results in a thick mitotic spindle. (A) Cells bearing a SET1 catalytic mutant allele (set1G951S) display a more robust mitotic spindle staining compared with wild-type cells (WT). Immunofluorescent confocal images of wild-type and set1G951S mitotic cells are shown. Tubulin (green) and DAPI (blue) stainings were used to identify cells undergoing mitosis in an asynchronous culture. Mitotic cells are outlined in white. (B) Cell cycle arrest and release experiments reveal that Pds1 is more stable in SET1 mutants (set1G951S) than in wild-type cells. Cells were arrested in G2/M and released into fresh medium. Samples were taken at the indicated time points, and whole-cell extracts and immunoblots were prepared and probed with α-Myc antibody to assess the protein levels of Myc-tagged Pds1. Immunoblots of Pgk1 served as a loading control. Cells from the indicated time points were taken for analysis by flow cytometry. Histograms of DNA content reveal the cell cycle profile of the indicated strains at specific times after cell cycle arrest and release. (C) Serial fivefold dilution assays of wild-type and mutants with the indicated genotype placed onto either control plates (YPD) or plates containing 30 µg/mL benomyl as in previous figures. (D) Immunofluorescent confocal images of wild type and mutants with the indicated genotypes. Staining and selection of cells were preformed as in A.
