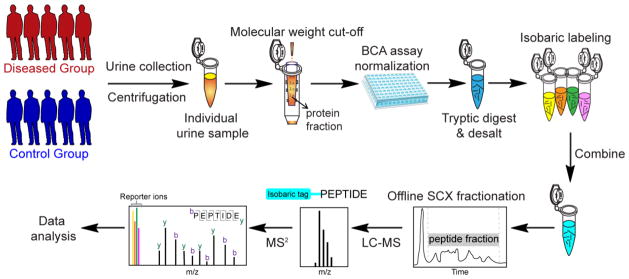
Fig. 3.
A representative workflow of quantitative urine proteomics using isobaric labeling. Urine samples are collected from human patients (control and disease groups) and centrifuged to remove particulates and cell debris. Protein fraction from each urine sample is obtained with molecular weight cut-off filter. Total protein concentration is measured with bicinchoninic acid (BCA) assay for normalization. After protein digestion and desalting, each sample is labeled by different channels of isobaric tags and combined after labeling reaction. Offline SCX LC is often performed to fractionate samples before LC-MS/MS analysis. The intensities of reporter ions are used for relative quantification.