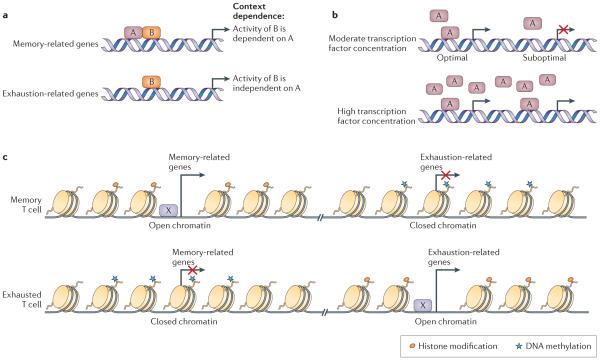
Figure 4. Transcriptional and epigenetic mechanisms of T cell exhaustion.
Altered usage of key transcription factors is associated with the altered transcriptional and developmental programme of T cell exhaustion. Several potential mechanisms exist. a | Different use of transcription factor binding partners is one potential mechanism for distinct context-dependent transcription factor activity. In the example shown, transcription factor B is closely associated with memory-related genes in the context of acute infection where transcription factor A is abundant (top panel). However, the same transcription factor B is linked with exhaustion-related genes in chronic infection (lower panel). Post-translational modifications of transcription factors and/or their subcellular localization may also be important in this setting. b | The dosage or concentration of a transcription factor could also provide a mechanism for context-dependent transcriptional function. Here, a transcription factor binds only at specific high-affinity binding sites if the amount of the factor is low (top panel). By contrast, at high transcription factor concentrations binding can occur more broadly (that is, occurring also at lower affinity sites), which leads to different transcriptional activity (lower panel). c | DNA methylation, histone modifications and the ‘chromatin landscape’ resulting from overall epigenetic regulation could provide a mechanism for context-specific transcription factor function in exhausted T cells (transcription factor X in this example). The enhancer landscape of a cell may determine how different transcription factors function.