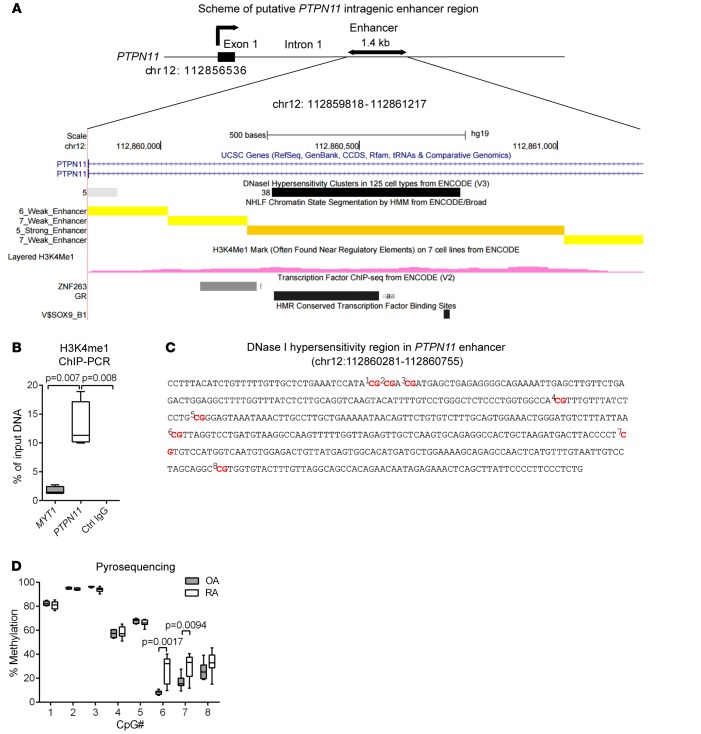
Figure 1. Identification of a putative intragenic PTPN11 enhancer displaying epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes versus osteoarthritis fibroblast-like synoviocytes.
(A) Scheme of the putative intragenic enhancer in chromosomal region 12q24.13, depicting an ENCODE-derived DNase I hypersensitivity cluster, chromatin state annotation tracks produced from ChromHMM, H3K4me1 marker, transcription factor ChIP sequencing, and human/mouse/rat conserved transcription factor–binding sites. Data were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/; assembly GRCh37/hg19). (B) After serum starving rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLS) for 24 hours, ChIP-PCR was performed using anti-H3K4me1 antibody on the PTPN11 enhancer or MYT1 exon 1 as a control or control (Ctrl) IgG on the PTPN11 enhancer. The occupancy of H3K4me1 or Ctrl IgG is shown as the percentage of input. Box-and-whisker plots depict median (line within box), 25th percentile and 75th percentile (bottom and top borders), and range of minimum to maximum values (whiskers) from 4 independent experiments with different RA FLS lines. Data were analyzed using the 2-tailed paired t test. (C) Sequence of DNase I hypersensitivity region in the PTPN11 enhancer (chr12:112860281-112860755) with 8 CpG dinucleotides. (D) Analysis of differentially methylated loci between RA (n = 8) and osteoarthritis (OA) (n = 6) FLS in the PTPN11 DNase I hypersensitivity region through pyrosequencing. Box-and-whisker plots depict the percentage of methylation of each CpG site. Data were analyzed using the 2-tailed unpaired t test with Welch’s correction.