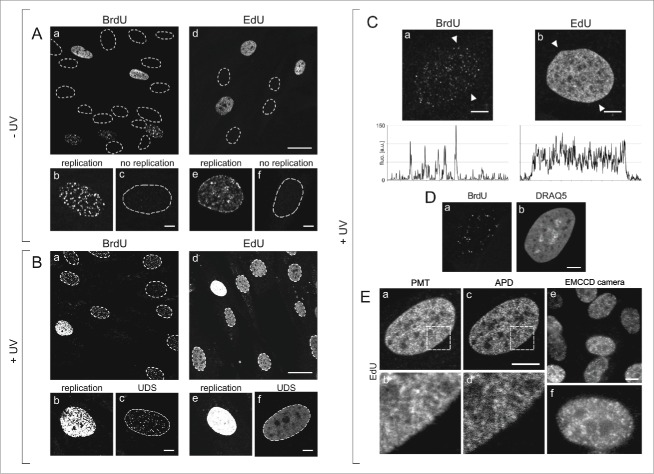
Figure 1.
Patterns of DNA replication and UDS revealed by incorporation of BrdU or EdU, and various modes of fluorescence detection. (A) Images of BrdU (a) or EdU (d) incorporated into DNA of replicating cells in asynchronous cell cultures, under standard conditions, prior to exposure to UVC. Contours of nuclei of non-replicating cells, based on transmitted light images (not shown), are marked with dotted lines. Panels below show magnified images of replicating and non-replicating nuclei, with incorporated BrdU (b, c) or EdU (e, f), showing typical replication patterns (b, e) and the absence of fluorescence signals in non-replicating cells (c, f). No EdU or BrdU signals were detected in non-replicating cells at low or high instrumental gain (data not shown). Scale bar 20 μm (a,d) and 5 μm (b,c,e,f). (B) Images showing BrdU (a) or EdU (d) in cells, recorded 2 h after exposure to UVC. The gain of the fluorescence detector (PMT) was set higher than in images in panel A, therefore the signals of replicating cells are oversaturated (b,e). Non-replicating cells show UDS signals of the incorporated BrdU or EdU (c,f). Non-replicating cells that do not activate UDS are also occasionally detected. Scale bar 20 μm (a,d) and 5 μm (b,c,e,f). (C) High magnification images showing BrdU (a) or EdU (b) incorporated in a process of UDS (UVC dose 10 J/m2), and demonstrating an apparent difference between the patterns revealed by incorporation of BrdU and EdU. Fluorescence profiles plotted along the lines marked with arrowheads are shown below each image. Scale bar 5 μm. (D) A pattern of UDS revealed by BrdU (a), distribution of dsDNA stained with DRAQ5 (b), and UDS against the background of dsDNA (c). Scale bar 5 μm. E. Patterns of UDS revealed by the incorporated EdU, detected by a standard photomultiplier in a confocal microscope (a,b), an avalanche photodiode (c,d), and a sensitive EMCCD camera in a widefield fluorescence microscope (e,f). Scale bar 10 μm.