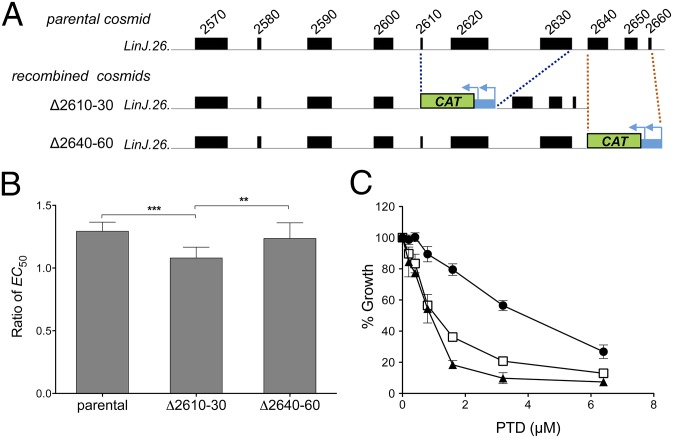
Fig. 5.
From cosmids to antileishmanial drug resistance genes. (A) Cosmid recombineering applied to cosmid LinJ.26b, which confers AMB resistance. Two segments of three consecutive genes at the 3′ end of cosmid LinJ.26b were independently replaced with a CAT marker, yielding recombined cosmids Δ2610–30 and Δ2640–60. Cosmid Δ2610–30 is deleted for the LinJ.26.2610, LinJ.26.2620, and LinJ.26.2630 genes, and cosmid Δ2640–60 is deleted for the LinJ.26.2640, LinJ.26.2650, and LinJ.26.2660 genes. (B) AMB EC50 ratio (candidate cosmid/empty vector) for L. infantum WT parasites transfected with the parental cosmid LinJ.26b or with its recombined derivative cosmids Δ2610–30 and Δ2640–60. Parasites transfected with empty cLHYG served as controls. Data are the mean ± SD of two independent experiments, each performed with five biological replicates and statistically analyzed using an unpaired two-tailed t test. **P ≤ 0.01; ***P ≤ 0.001. (C) Effect of PTD on the growth kinetics of L. infantum WT parasites transfected with cosmid LinJ.31b (circle) and its PRP1-null derivative generated by cosmid recombineering (triangle). The same strategy used for PTR1 deletion (Fig. 4A) was used here for PRP1. Parasites transfected with empty cLHYG served as controls (square). Data are the mean ± SD of three biological replicates.