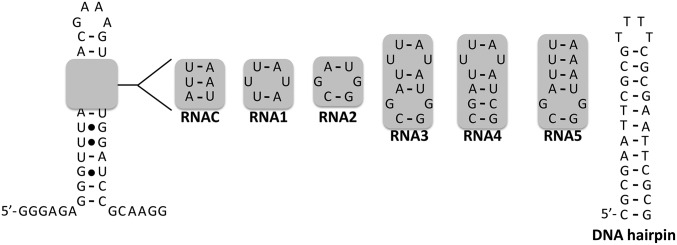
Fig. 3.
Secondary structures of the nucleic acids to which the binding of monomeric small molecules 1 and 2 and dimeric compound 3 was assessed. RNAC is a control RNA that does not contain target sites for 1 or 2 and has a single-nucleotide change relative to RNA1. RNA1 contains the motif present in the Drosha processing site of pri-miR-96. RNA2 contains the 1 × 1 nt GG internal loop that is adjacent to the Drosha site. RNA3 contains both the Drosha site and the adjacent 1 × 1 nt GG internal loop. RNA4 contains the UU internal loop present in the Drosha site and a single-nucleotide change to mutate the 1 × 1 nt GG internal loop to a GC pair. RNA5 contains the GG internal loop, but the UU loop has been mutated to an AU pair.