Fig. 1.
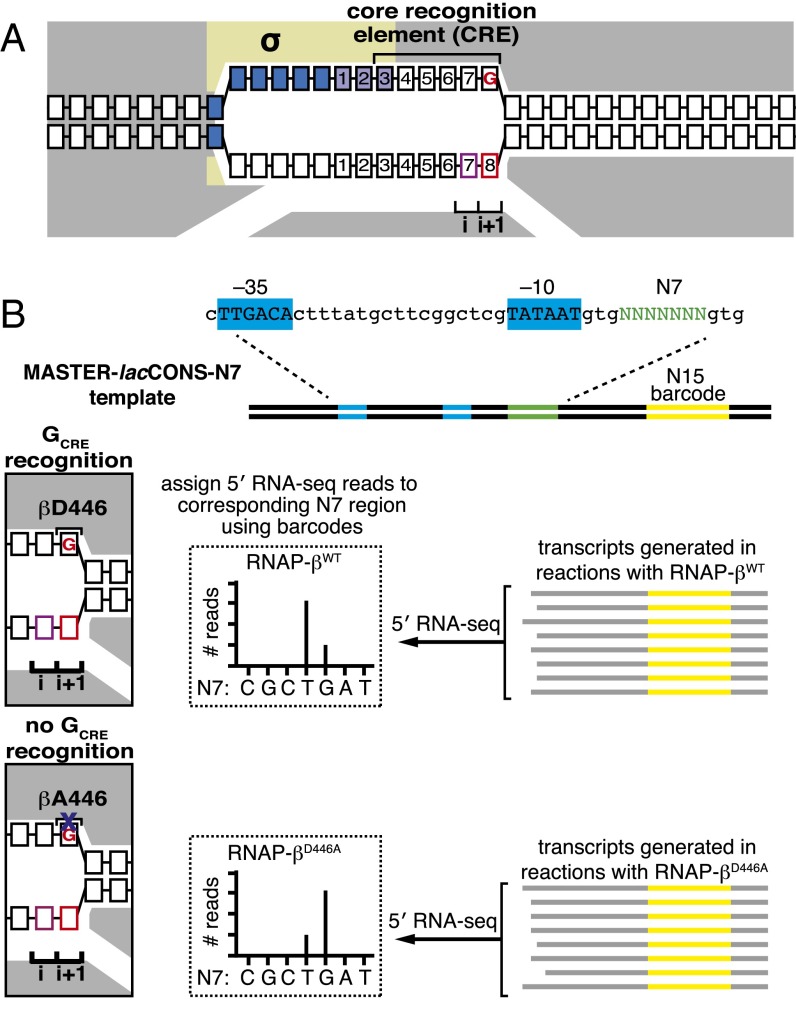
Analysis of effects of sequence-specific RNAP–CRE interactions by MASTER (11). (A) RPo for TSS at position 7. Gray, RNAP; yellow, σ; blue boxes, −10 element nucleotides; purple boxes, discriminator nucleotides; black boxes, DNA nucleotides (non–template-strand nucleotides above template-strand nucleotides; nucleotides downstream of −10 element numbered); pink box, TSST; red box, TSS+1T; i and i+1, RNAP active-center initiating NTP binding site and extending NTP binding site; red “G,” GCRE. (B, Upper) DNA fragment carrying the MASTER template library lacCONS-N7. Promoter −35 and −10 elements are indicated. Randomized nucleotides are green and 15-nt barcode sequence in the transcribed region is yellow. (Lower Right) 5′ RNA-seq analysis of RNA products generated from the MASTER-N7 template library in vitro. The sequence of the barcode is used to assign the RNA product to an N7 region and the sequence of the 5′ end is used to define the TSS. (Lower Left) Structural organization of downstream end of transcription bubble in RPo for promoter containing GCRE formed with WT RNAP (Upper) or RNAP derivative carrying the βD446A substitution (Lower). Black “βD446,” RNAP β-subunit residue that makes sequence-specific favorable interaction with GCRE. Black “βA446,” RNAP β-subunit residue in mutant RNAP defective in sequence-specific interaction with GCRE. Other rendering and colors as in A.