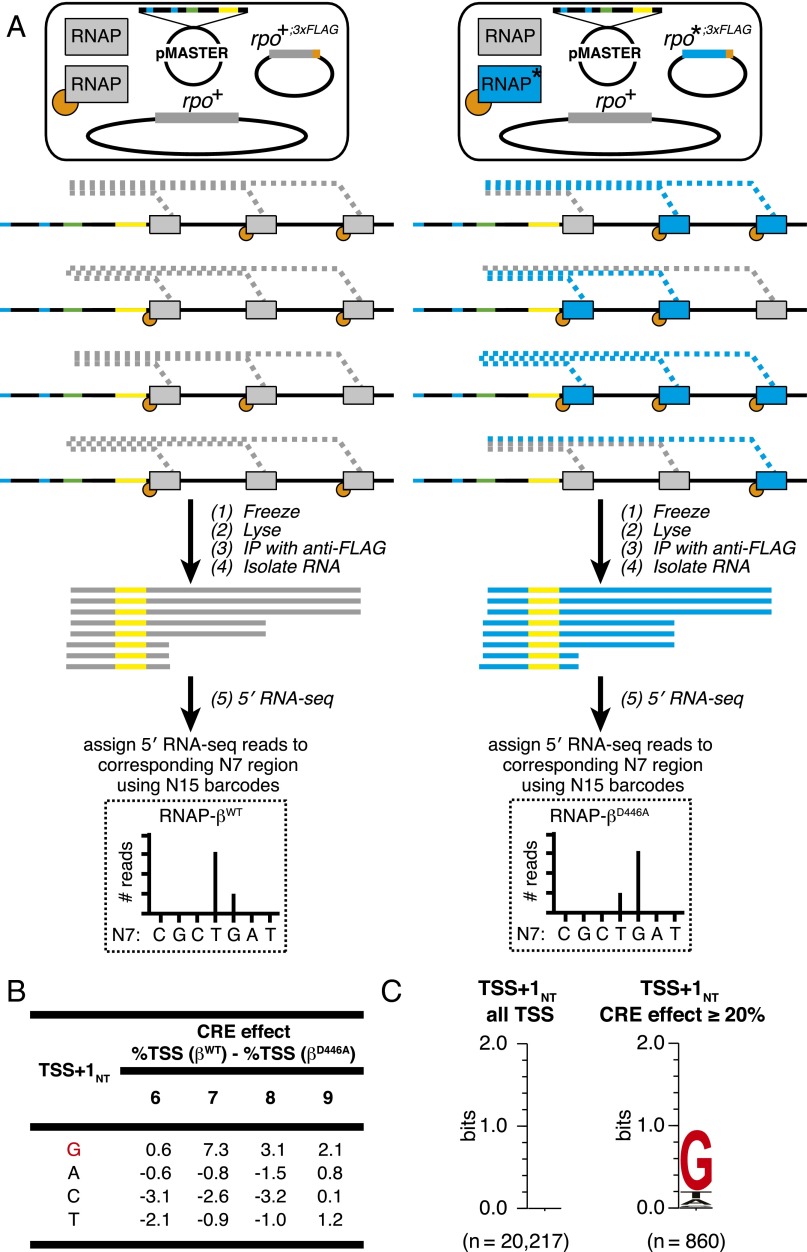
Fig. 4.
Effects of disrupting RNAP–GCRE interactions in vivo: 5′ mNET-seq analysis of 47 (∼16,000) consensus promoter derivatives. (A) Steps in 5′ mNET-seq analysis of TSS selection from plasmid-borne MASTER template library: (Top) RNAP derivatives in cells (the blue RNAP derivative with asterisk is RNAP-βD446A); (Middle) RNAPs on the same MASTER template in four cells (RNA products in blue are associated with RNAP-βD446A); and (Bottom) isolation of RNA products after immunoprecipitation with anti-FLAG affinity gel and sequencing analysis of RNA 5′ ends. In this example, TSS selection at the T in the middle of the randomized TSS region is decreased with the mutant RNAP derivative. (B) Effect of sequence at TSS+1NT on %TSS for RNAP-βWT vs. RNAP-βD446A. Table lists the difference in %TSS (%TSS for RNAP-βWT − %TSS for RNAP-βD446A) at positions 6, 7, 8, or 9 for TSS-regions carrying G, A, C, or T at TSS+1NT. (C) Sequence preferences for TSS+1NT. Sequence logo (33) for TSS+1NT of above-threshold TSS positions located 6–9 bp downstream of the −10 element (Left) and TSS positions located 6–9 bp downstream of the −10 element that exhibited a large, ≥20%, reduction in %TSS in 5′ mNET-seq analysis of RNAP-βD446A vs. 5′ mNET-seq analysis of RNAP-βWT (Right).