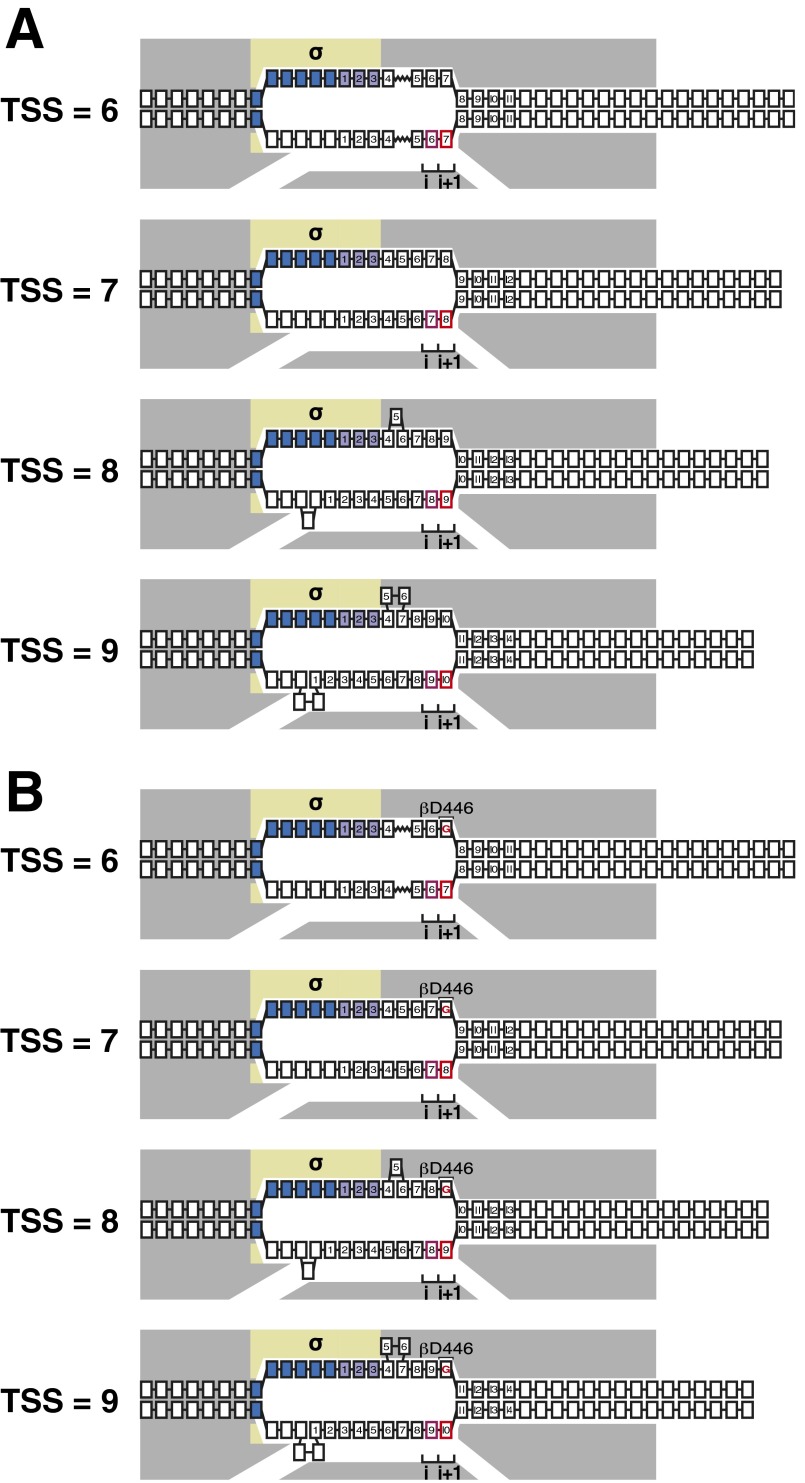
Fig. S1.
Model for TSS selection and hypothesis for effects of RNAP–CRE interactions on TSS selection. (A) Model for TSS selection: changes in TSS selection result from changes in DNA scrunching and antiscrunching in RPo (11–13). TSS = 6 through TSS = 9, RPo engaged in TSS selection at positions 6 through 9 nt downstream of −10 element; gray, RNAP; yellow, σ; blue boxes, −10 element nucleotides; purple boxes, discriminator nucleotides; black boxes, DNA nucleotides (non–template-strand nucleotides above template-strand nucleotides; nucleotides downstream of −10 element numbered); pink box, TSST; red box, TSS+1T; i and i+1, RNAP active-center initiating NTP binding site and extending NTP binding site. Scrunching is indicated by bulged-out nucleotides. Antiscrunching is indicated as a stretched nucleotide–nucleotide linkage. Exact positions and conformations of scrunched nucleotides of the nontemplate and template DNA strands remain to be determined. (B) Hypothesis for effects of RNAP–CRE interactions on TSS selection: RNAP–CRE interactions modulate TSS selection by modulating the extent of scrunching and antiscrunching in RPo. TSS = 6 through TSS = 9, RPo engaged in TSS selection at positions 6 through 9 nt downstream of −10 element at promoters that contain GCRE (i.e., promoters that contain G at the non–template-strand position opposite TSS+1T). Red “G,” GCRE. Black “βD446,” RNAP β-subunit residue that makes sequence-specific favorable interaction with GCRE. Other rendering and colors as in A.