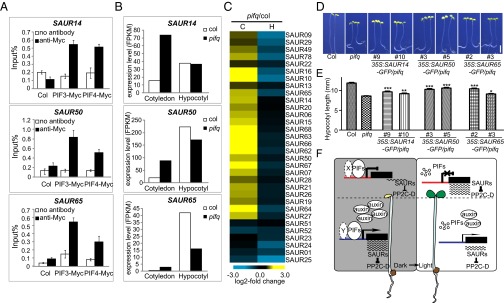
Fig. 4.
The regulation of lirSAUR gene expression by PIFs and a working model of how light differentially regulates the development of cotyledons and hypocotyls. (A) ChIP-qPCR analysis of 4-d-old dark-grown seedlings showing specific enrichment of PIF3 and PIF4 on SAUR14, SAUR50, and SAUR65 promoters. Col and no antibody were used as controls. (B) Expression of SAUR14, SAUR50, and SAUR65 in the cotyledons and hypocotyls of 4-d-old dark-grown pifq and WT seedlings. (C) Heat map of lirSAURs in cotyledons (C) and hypocotyls (H) of 4-d-old dark-grown pifq seedlings compared with WT (pifq/col) by RNA-seq analysis. (D) Representative 4-d-old dark-grown seedlings of WT, pifq, and SAUR14, SAUR50, and SAUR65 overexpression transgenic lines in pifq background. (Scale bar: 2 mm.) (E) Hypocotyl lengths of the seedlings shown in D. The data are shown as mean ± SEM (n > 20). Statistical significance was calculated using Student’s t test compared with pifq. ***P < 0.001, **P < 0.01, *P < 0.05. (F) A working model of how light differentially regulates the growth of cotyledons and hypocotyls during deetiolation. X and Y represent possible different PIF cofactor(s) in cotyledons and hypocotyls, respectively. Promoters are labeled in two colors (red in the cotyledons and blue in the hypocotyls) indicating the possible differences in epigenetic modifications of the same lirSAUR gene in these two organs.