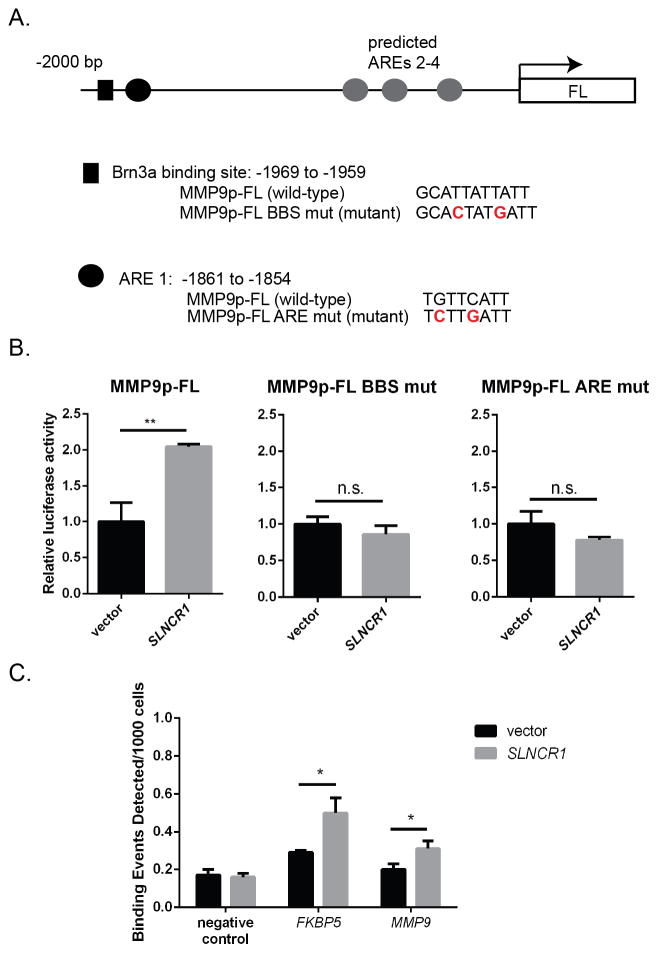
Figure 6. AR and Brn3a binding sites are required for SLNCR1-induced upregulation of the MMP9 promoter.
(A) Schematic presentation of the 2 KB MMP9 promoter cloned upstream of the firefly luciferase reporter. The black box denotes a predicted Brn3a binding site. The wild-type and mutated sequences are shown below. The black circle denotes a functional ARE, with wild-type and mutated sequences below. The grey circles denote additional predicted AREs. (B) Mutation of either the Brn3a binding site (MMP9p-FL BBS mut) or the ARE (MMP9p-FL ARE mut) prevents SLNCR1-mediated upregulation of the MMP9 promoter. Assay was completed as in Figure 3G. Error bars represent standard deviation from four reactions. (C) AR-ChIP from A375 cells transfected with either vector or SLNCR1-expressing plasmid. qPCR was performed using primers specific to the regions indicated, including primers corresponding to a gene desert (negative control). * p-value < 0.05, ** p-value < 0.005, n.s. = not significant. See also Figure S6.