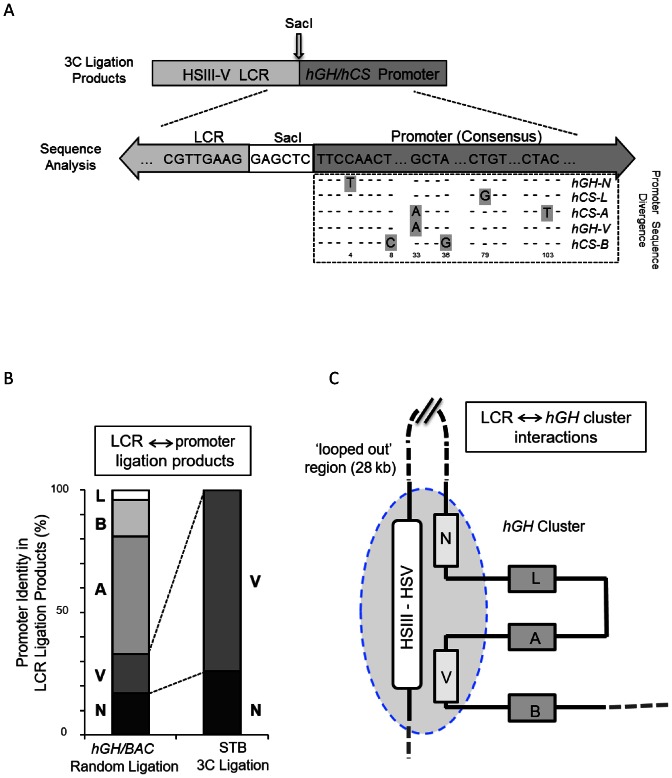
Figure 2.
The hGH LCR (HSIII–V) selectively loops to the hGH-N and hGH-V promoters in primary human placental STBs. (A) Approach to specifying interactions of the hGH LCR with individual hGH/hCS promoters. 3C-ligation products generated between the SacI fragments containing the hGH LCR (anchor) and the gene promoter (Pro) fragments in the random ligation control (SacI-digested hGH/BAC plasmid) and in STB chromatin (as in Figure 1) were cloned and individually sequenced. The identities of the LCR anchor fragments and SacI recognition sequence (GAGCTC) were confirmed in each ligation product and the specific identity of each linked promoter fragment was determined via unique distinguishing sequences (as highlighted in the promoter sequence alignment). (B) The LCR interacts specifically with the hGH-N and hGH-V promoters. The relative distributions of each of the five gene promoters ligated to the placental LCR (as determined in A, above) from the random ligation of SacI-digested hGH/BAC DNA and from the 3C analysis of human placental STB chromatin are displayed as indicated. These data represent the sequence analysis of 96 and 104 subcloned ligation products, respectively. (C) Schematic of interactions between the hGH LCR and the hGH gene cluster in primary human STB cells. This model is based on the 3C analysis using the LCR anchor primer. The specificity of promoter identities (as in A) and their relative frequencies (as in B) are represented with HSIII–V LCR being situated in proximity to the hGH-N and hGH-V promoters. The 28 kb region between the hGH LCR and the hGH-N promoter lacks interactions with the LCR and is therefore represented as a ‘looped-out’ segment. The hCS-L, hCS-A and hCS-B genes are all excluded from LCR interactions and are similarly shown as ‘looped-out’ from the hub of LCR interactions with the two GH genes.