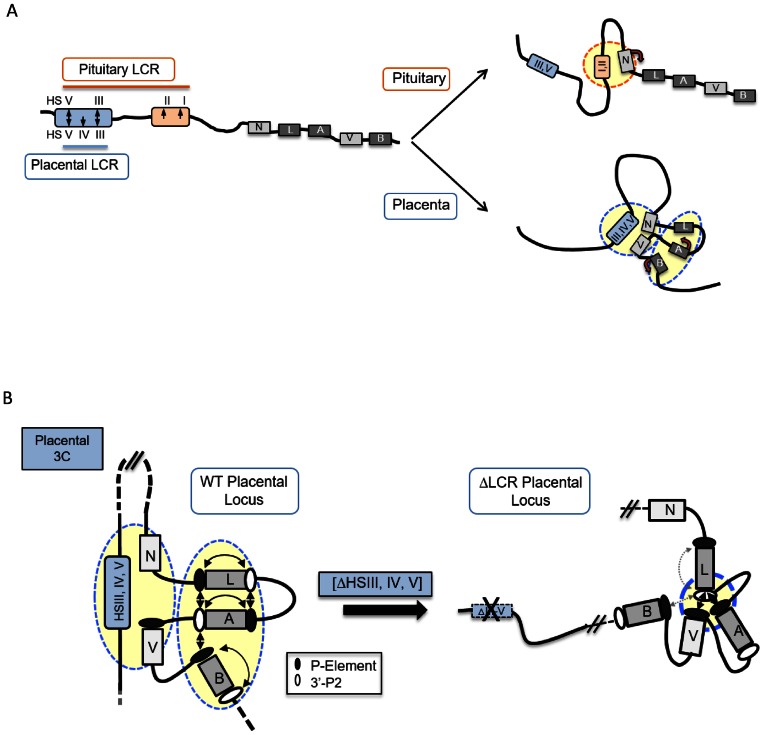
Figure 7.
3D chromatin architectures at the hGH multigene locus in the pituitary and placenta. (A) Distinct patterns of long-range LCR looping in the pituitary and placenta parallel tissue-specific gene expression from the hGH gene cluster in the two tissues. During the activation of pituitary hGH locus, the pituitary-specific HSI enhancer directly interacts with its target hGH-N promoter via chromatin looping (14). In contrast, in the placenta the HSIII–V LCR region loops to two hGH promoters, displacing the activated hCS genes into a tightly packed ‘hCS chromatin hub’. (B) The organization of hGH gene cluster in placental chromatin is driven by long-range LCR interactions. HSIII–V LCR interactions segregate the hGH-N and hGH-V genes from the hCS gene hub within the intact hGH locus in the placenta. Deletion of the placental LCR (HSIII–V) triggers a dramatic reconfiguration within the cluster that disrupts the ‘hCS chromatin hub’ with a corresponding loss of hCS transcriptional control.