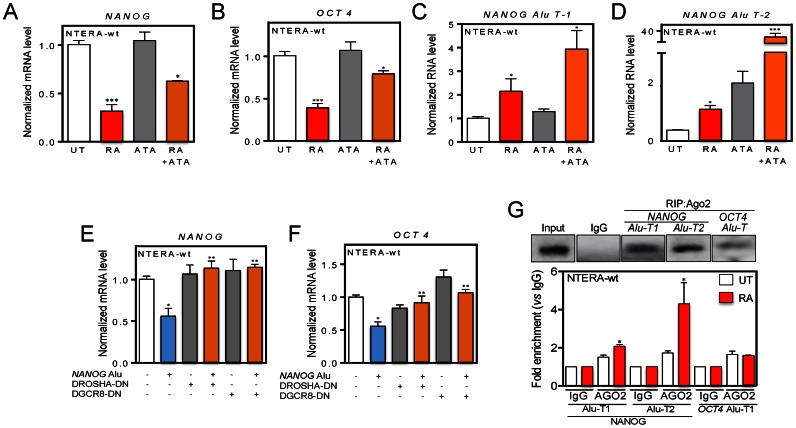
Figure 8.
RISC inhibition and microprocessor knockdown rescues NANOG and OCT4 expression likely through loading of NANOG Alu-derived transcripts into AGO2. (A and B) NTERA-wt cells were left untreated (UT), treated with 25 μM ATA (aurintricarboxylic acid) or with 1 μM RA + 25 μM ATA and the expression of NANOG (A) and OCT4 (B) determined as above. (C and D) NTERA-wt cells were left untreated (UT), treated with 25 μM ATA or with 1 μM RA + 25 μM ATA and the expression of NANOG Alu-T1 (C) and Alu-T2 (D) determined by RT-qPCR. The oligonucleotides used are indicated in Supplementary Table S1. (E and F) The NANOG Alu was transfected in NTERA-wt cells with or without dominant-negative forms of DROSHA or DGCR8 and their effects on NANOG (E) and OCT4 (F) expression analyzed by RT-qPCR. (G) RNA was extracted from NTERA-wt cells and RNA immunoprecipitations (RIP) for NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts performed using an anti-AGO2 antibody. RIP was quantified by qPCR using the oligonucleotides indicated in Supplementary Table S1. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A and B: n = 6, two technical replicates in three biological replicates; panels C and D: n = 6, two technical replicates in three biological replicates; panels E and F: n = 4, two technical replicates in two biological replicates; panel G: n = 4, two technical replicates in two biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.