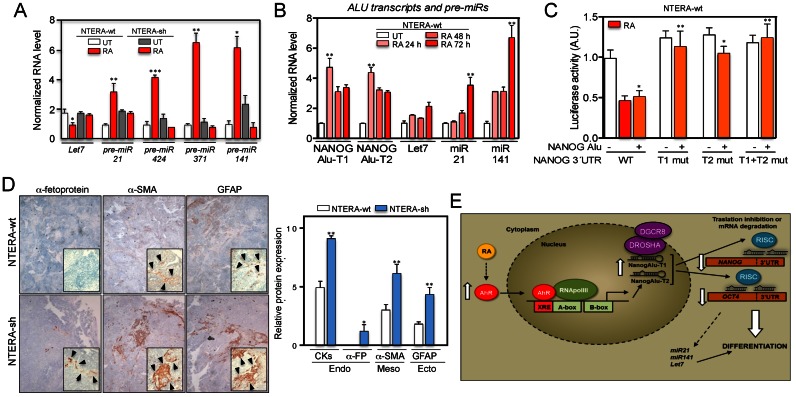
Figure 9.
NANOG Alu-derived transcripts could act through a miRNA mechanism to control NANOG and OCT4 repression during differentiation. (A) NTERA-wt and NTERA-sh cells were left untreated (UT) or treated with 1 μM RA for 48 h and the levels of pre-miRs pre-Let7, pre-miR-21, pre-miR-424, pre-miR-371, pre-miR-141 quantified by RT-qPCR using the oligonucleotides indicated in Supplementary Table S1. (B) Both cell lines were treated with 1 μM RA for the indicated times and the levels of NANOG Alu-T1 and Alu-T2, pre-Let7, pre-miR-21 and pre-miR-141 determined by RT-qPCR. (C) A NANOG 3′UTR-Luc construct and their deletion mutants lacking the homology regions for NANOG Alu-T1 (T1 mut), Alu-T2 (T2 mut) or both (T1 + T2 mut) were co-transfected with the NANOG Alu in NTERA-wt cells. Firefly luciferase activity was measured and normalized by renilla luciferase. Control experiments were done in presence of RA (dashed). (D) NTERA-wt and NTERA-sh cells were injected subcutaneously in immunodeficient Swiss nude mice (Charles River) and the tumors formed collected and analyzed by immunohistochemistry using endodermal (pan-cytokeratins and α-fetoprotein), mesodermal (α-smooth muscle actin) and ectodermal (glial fibrilar acidic protein, GFAP) markers. Immunohistochemistry sections were analyzed blinded by eight independent observers and protein expression quantified in a 0–10 scale. Arrowheads indicate positivity areas used to estimate protein expression. (E) Scheme of the mechanism proposed for the Alu-dependent repression of NANOG and OCT4 during carcinoma cell differentiation. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A and B: n = 6, three technical replicates in two biological replicates; panel C: n = 4, two technical replicates in two biological replicates; panel D: n = 4 mice for each cell line. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.