Figure 6.
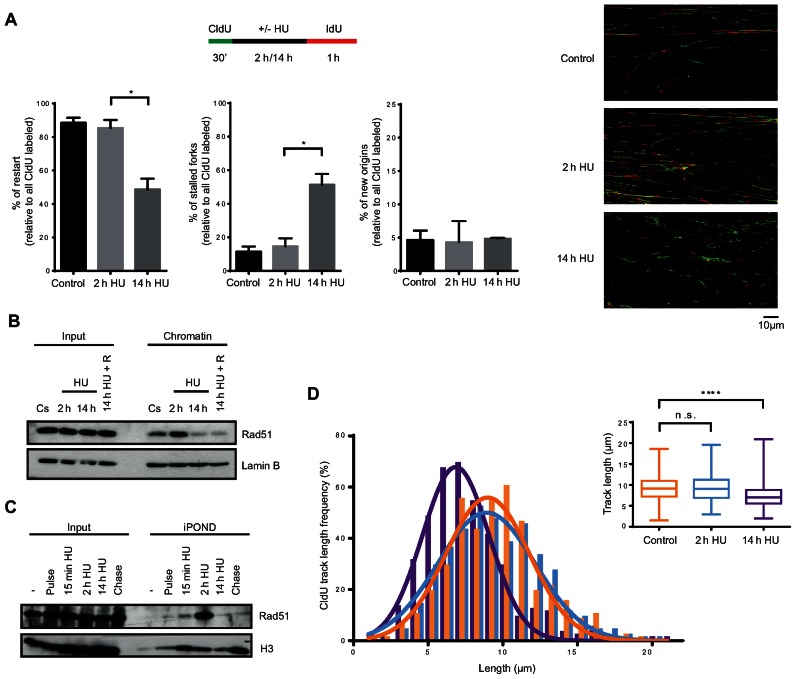
Replication forks of hTERT-RPE cells become inactivated after prolonged DNA replication inhibition. (A) Labelling protocol for DNA fibre analysis (upper panel). S-phase synchronized (by single thymidine block) hTERT-RPE cells were treated as indicated and then DNA fibres were prepared and labelled with anti-BrdU antibodies. The percentage of replication fork restart, stalled replication forks and new origin firing relative to total CldU labelled fibres are shown in the graphs. At least 1500 fibres from three independent experiments were counted in each condition. Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, *P < 0.05). Representative images of each condition are shown (right panel). (B) S-phase synchronized hTERT-RPE cells were treated during the indicated time with HU or left untreated (Cs). Cells were harvested just after the treatment or after a 30 min release into fresh media (14 h HU + R). Chromatin extracts were prepared and analysed by western blot with the indicated antibodies. Input: whole cell lysates. Lamin B was used as loading control. (C) hTERT-RPE cells were synchronized in S-phase and labelled with EdU during 15 min before treating them with HU during the indicated time. EdU was present in the media for additional 15 min in HU-treated cells. Isolated proteins on nascent DNA (iPOND) were analysed by western blot with the indicated antibodies. Input: nuclear extracts. Pulse: cells were harvested just after EdU labelling. Chase: 15 min EdU + 2 h thymidine (50 μM) chase. (-): no EdU. (D) DNA fibres from (A) were used to measure the CldU track length. Three hundred fibres were counted in each condition. CldU track length distribution and statistical analysis of CldU track distribution are shown. Box and whiskers show Min, Max, Median and first quartiles. Values marked with asterisks are significantly different (unpaired t-test, n.s.: non-statistically significant, ****P < 0.0001).