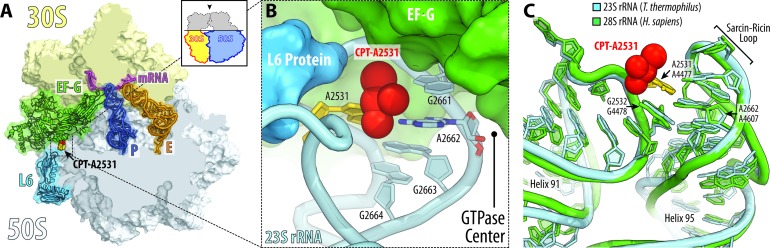
Figure 6.
Cisplatin modification site near the GTPase activating center of the ribosome. (A) Overview and (B) close-up view of the cisplatin position in the vicinity of the ribosome GTPase activating center. The structure of cisplatin-modified ribosome is superimposed with the structure of ribosome-bound EF-G (green) (PDB ID: 4V5F (44)). Superposition is based on the alignment of the 23S rRNA. 30S subunit is shown in light yellow, 50S subunit is in light blue. mRNA is shown in magenta and tRNAs are displayed in dark blue for the P site and in orange for the E site. tRNA for the A site is omitted for clarity. The view in (A) is from the top after removing the head of the 30S subunit and protuberances of the 50S subunit, as indicated by the inset. Note that cisplatin moiety of A2531 lies at the ribosome/EF-G interface in the immediate vicinity of the GTPase activating center (nucleotide A2662). (C) Superposition of the 23S rRNA from Thermus thermophilus (light blue) in the vicinity of the GTPase activating center with the homologous region of the 28S rRNA from Homo sapiens (green; PDB ID: 4V6X (43)). Note that due to extremely high conservation of the GTPase activating center, human ribosomes are likely to be modified by cisplatin at the same site as bacterial ribosomes.