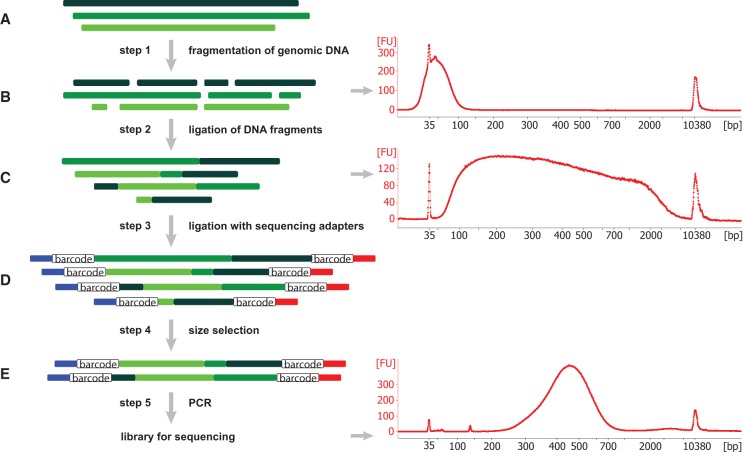
Figure 1.
Schematic of the SMASH method and size analysis. (A) Three representative genomic DNA molecules, shown in different shades of green, originate from different chromosomes or distant regions of the same chromosome. (B) By sonication and restriction enzyme cleavage, these molecules are fragmented into short double-stranded DNA fragments with average length of 40–50 bp, as shown in the Bioanalyzer result at right. (C) These short DNA pieces are then partially end-repaired and combined into longer stretches of DNA with lengths ranging from 50 bp to 7 kb. Consequently, each resulting chimeric DNA molecule contains short DNA fragments from different locations (shown by varying colors). (D) These DNA stretches are ligated to sequencing adaptors containing sample barcodes, shown in blue and red lines, with the open box designating the sample barcodes. (E) Size selection is carried out to enrich for DNA fragments in the size range of 250–700 bp, which is confirmed via Bioanalyzer. After final PCR, libraries are ready for sequencing. “FU” in the Bioanalyzer plots refers to relative fluorescence units.