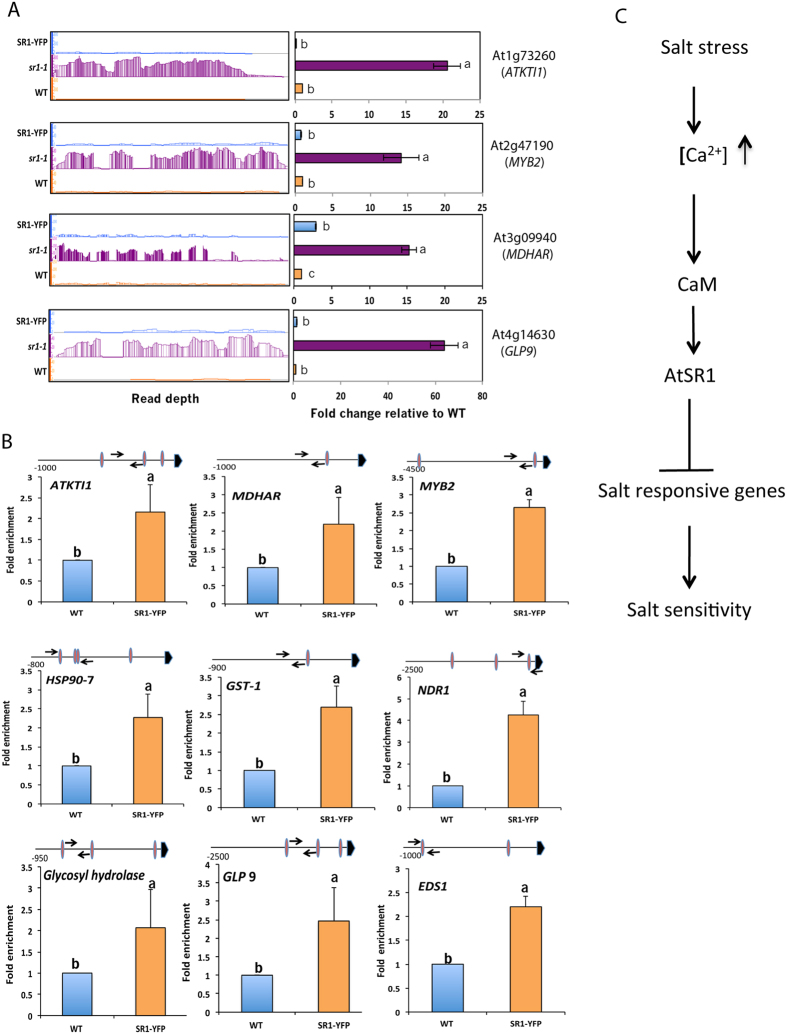
Figure 7. SR1 regulation of salt-responsive genes.
(A) Expression levels of a few representative salt stress-responsive genes in WT, sr1-1 and SR1-YFP. Left Panels: relative sequence read abundance (IGB view) as histograms in wild type (WT), SR1 mutant (sr1-1) mutant and the complemented line (SR1-YFP). The Y-axis indicates read depth with the same scale for all three lines. Right panels: Expression analysis of salt-responsive genes using RT-qPCR. Panels on right show fold change in expression level relative to WT. WT values were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. The error bars represent SD. (B) ChIP-PCR of upstream regions of salt-responsive genes containing VCGCGB or MCGTGT or MCGCGT + VCGCGB. Chromatin from 15-day-old seedlings from WT and SR1-YFP was immunoprecipitated with anti-GFP antibody and used in PCR with primers flanking the putative SR1 binding sites. The results obtained from four independent ChIP experiments were used to calculate fold enrichment. Data was normalized to DNA input levels as well as ACTIN2. The values of WT were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. Schematic diagram over each panel shows SR1 binding sites (as oval shape) and the location of primers used in ChIP-PCR are indicated with arrows. Bold arrowhead indicates TSS. (C) Proposed model for the role of SR1 in salt stress response (see text for details).