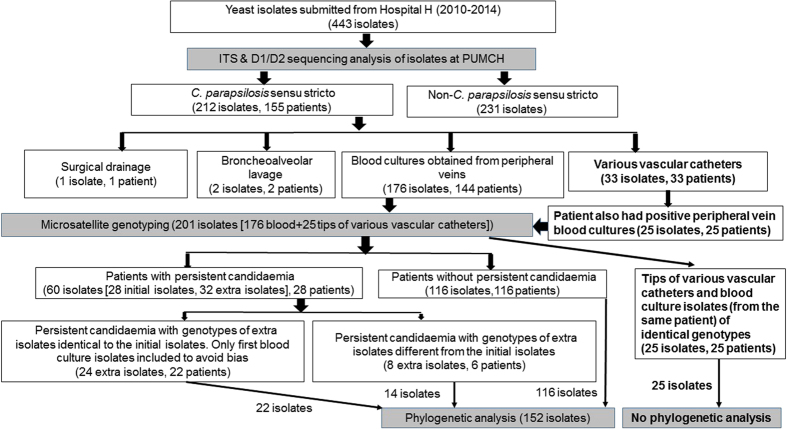
Figure 1. Flowchart of species identification, microsatellite genotyping, and phylogenetic analysis of yeast isolates collected from Hospital H in the CHIF-NET programme from 2010 to 2014.
All the 443 yeast isolates collected from Hospital H were further identified to species levels by using sequencing analysis of the internal transcribed spacer region and D1/D2 domain of the 26S rRNA gene at PUMCH. The 33 isolates of Candida parapsilosis sensu stricto yielded from tip cultures of various vascular catheters were collected from 33 patients. Among the 33 patients, 25 had positive culture of same yeast species from tip of various vascular catheters as well as from blood samples collected from peripheral veins. Microsatellite genotyping was also performed for the 25 isolates from various vascular catheter tips to demonstrate the possibility of catheter-related candidaemia. Genotypes of isolates recovered from various vascular catheter tips and peripheral veins in each of the 25 patients were identical. Among the 60 C. parapsilosis sensu stricto isolates recovered from 28 patients with persistent candidaemia (32 extra isolates in addition to the 28 initial blood isolates), 24 were recovered from 22 patients (two patients with three episodes of candidaemia [2 extra isolates]) with persistent candidaemia showed similar genotypes as the initial culture isolates. Thus to avoid bias, only the 22 first blood culture isolates of the 22 patients were included further phylogenetic analysis. The remaining eight C. parapsilosis sensu stricto isolates from six patients with persistent candidaemia (two patients with three episodes of candidaemia [2 extra isolates]) exhibited different genotypes to those of the initial blood culture isolates. These 8 extra isolates and the six first isolates of the six patients (n = 14) were all included in the phylogenetic analysis. As a result, the final number of C. parapsilosis sensu stricto isolates subjected to microsatellite genotyping and phylogenetic analysis was 201 and 152, respectively.