Figure 1. CHRNA7 genomic structure and linkage disequilibrium (LD) plot.
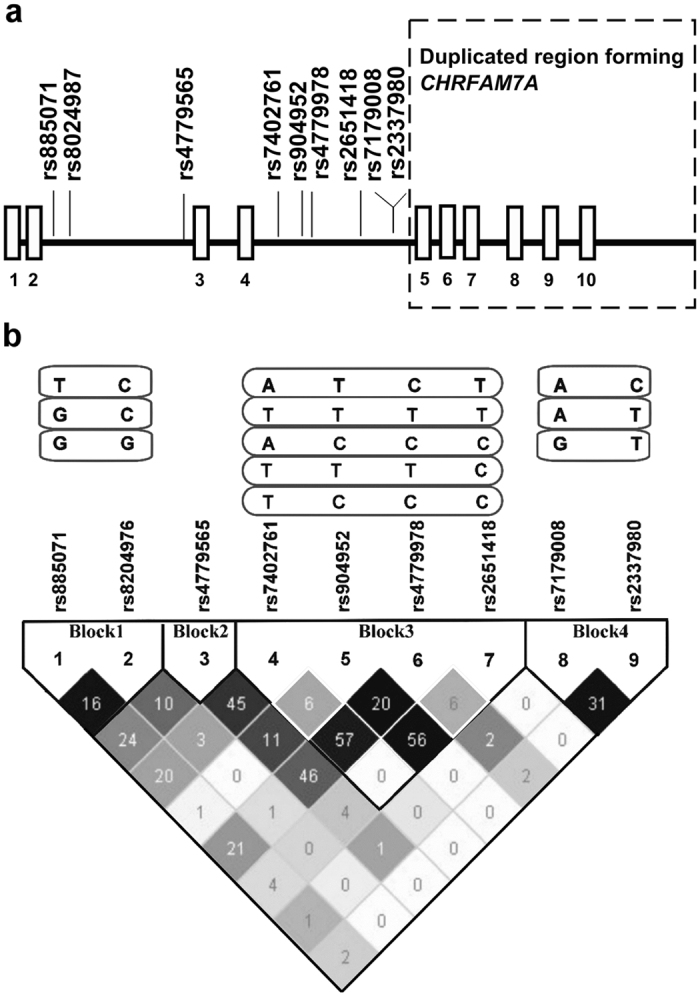
(a) shows the genomic structure of the CHRNA7 gene and the htSNPs in this study. The dashed rectangle indicates the region duplicated in the fusion gene, CHRFAM7A. Exons and introns are indicated by boxes and lines, respectively (not to scale). (b) shows the LD plot generated by Haploview program using genotype data from this study. Levels of pairwise D′, which indicate the degree of LD between two htSNPs, are shown in the LD structure in gray scale. Levels of pairwise r2, which indicate the degree of correlation between two SNPs, are shown as the number in each cell. Common (frequency ≥5%) haplotypes were identified in each haplotype block. A modified Gabriel et al. algorithm was used to define the haplotype block.
