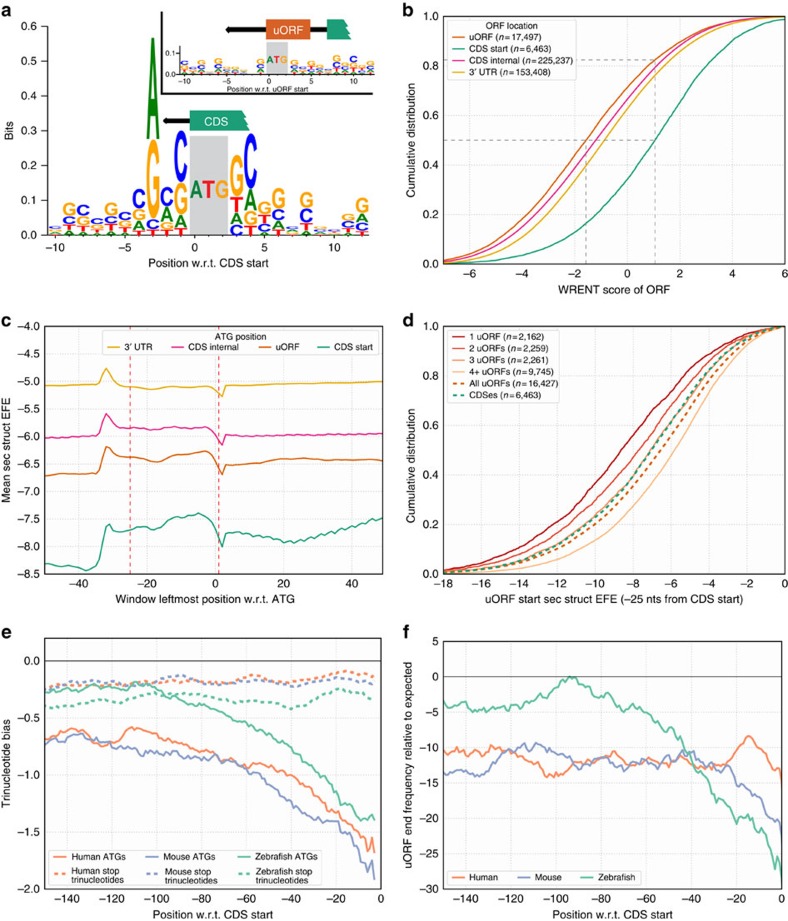
Figure 1. uORF sequence features are associated with weak repressiveness.
(a,b) Analysis of initiation context sequence in mouse. (a) Sequence motif over CDS starts (±10 nucleotides around the annotated AUG start) constructed from CDS TE-weighted position-specific scoring matrices (PSSMs) of coding transcripts lacking uORFs. Height on vertical axis represents weighted relative entropy (WRENT) at individual positions. Inset shows sequence motif at uORF starts, using uORF TE-weighted PSSMs of coding transcripts with one non-overlapping uORF. (b) Cumulative distribution of WRENT scores around AUGs at various positions in coding transcripts. Dotted lines indicate median uORF and CDS WRENT scores, as well as the proportion of uORFs (∼83%) with WRENT scores less than the median CDS WRENT score. (c,d) Analysis of initiation context secondary structure in mouse. (c) Meta-profiles of predicted secondary structure ensemble free energies (EFEs; sliding 35-nucleotide window) around AUGs in 5′ leaders, CDSes, and 3′ UTRs. A more negative EFE indicates more stable secondary structure. Red dotted lines indicate the positions −25 and +1 from ORF start that were used for subsequent analyses. (d) Cumulative distribution plot of initiation context secondary structure (at position +1 from the ORF start) of uORFs in transcripts with varying number of uORFs. Distributions of secondary structure EFEs for all uORFs and for CDS initiation contexts are indicated (dashed lines). (e,f) Distribution of uORFs. (e) ATG (that is, start codon; solid lines) and stop codon (dashed lines) moving average of positional trinucleotide biases in 5′ leaders are plotted against their position with respect to the CDS start, for all 3 vertebrates. ATGs, but not stop codon trinucleotides are specifically depleted near the CDS start. (f) Depletion in the frequency of uORF ends (moving average over 24 nucleotides) relative to expected frequencies from shuffled 5′ leaders, plotted against uORF-end position with respect to CDS, for all 3 vertebrates. uORF ends are specifically depleted in the 5′ leader near the CDS, most significantly in zebrafish and mouse transcripts, less so in human transcripts.