Figure 1. Targeted phenotypic culturing facilitates bacterial discovery from healthy human faecal microbiota.
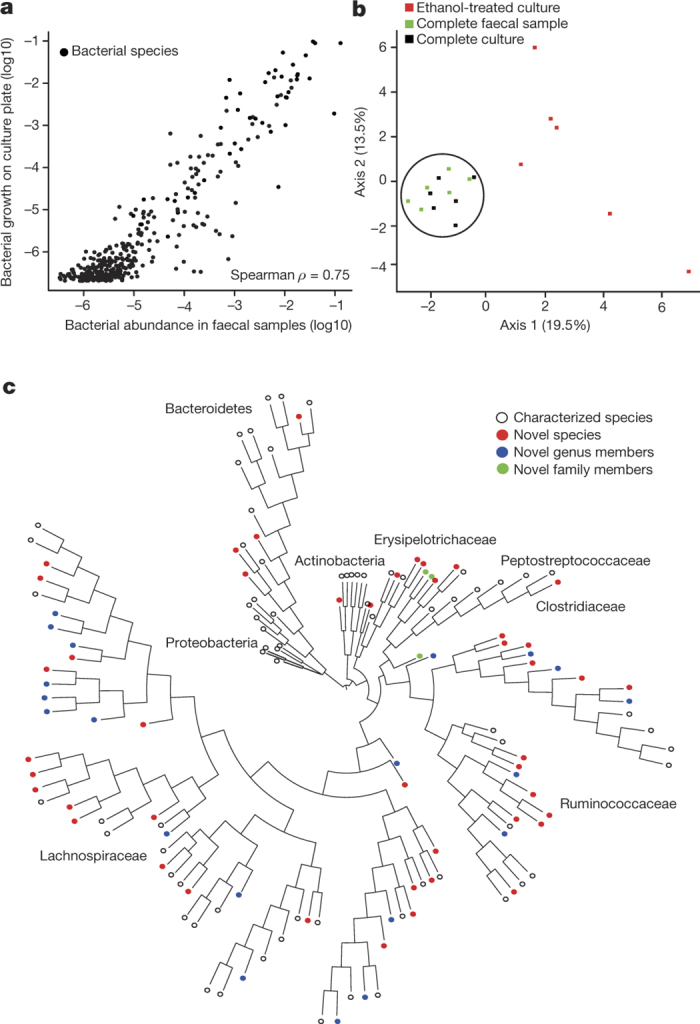
a, Relative abundance of bacteria in faecal samples (x axis) compared with relative abundance of bacteria growing on YCFA agar plates (y axis) as determined by metagenomic sequencing. Bacteria grown on YCFA agar are representative of the complete faecal samples as indicated by Spearman ρ = 0.75 (n = 6). b, Principal component analysis plot of 16S rRNA gene sequences detected from six donor faecal samples (n = 6), representing bacteria in complete faecal samples (green), faecal bacterial colonies recovered from YCFA agar plates without ethanol pre-treatment (black) or with ethanol pre-treatment to select for ethanol-resistant spore-forming bacteria (red). Culturing without ethanol selection is representative of the complete faecal sample, ethanol treatment shifts the profile, enriching for ethanol-resistant spore-forming bacteria and allowing their subsequent isolation. c, Phylogenetic tree of bacteria cultured from the six donors constructed from full-length 16S rRNA gene sequences. Novel candidate species (red), genera (blue) and families (green) are shown by dot colours. Major phyla and family names are indicated. Proteobacteria were not cultured, but are included for context.
