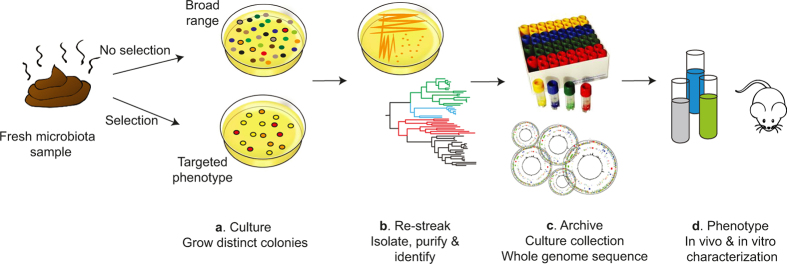
Extended Data Figure 1. A workflow for culturing, archiving and characterization of the intestinal microbiota.
a–d, Schematic diagram of the workflow, encompassing bacterial culturing and genomics to isolate and characterize bacterial species from the human intestinal microbiota. The process incorporates several steps, which are culture, re-streak, archive and phenotype. a, Fresh faecal samples are left untreated or are treated to select for bacteria with a desired phenotype (such as sporulation). The stool is homogenized and then serially diluted and then aliquots of the homogenate are inoculated on YCFA agar to culture bacteria. b, Isolates are identified by selecting single colonies that are streaked to purity and full-length 16S rRNA genes are amplified and sequenced. c, Each unique, novel and desired isolate is archived frozen in a culture collection and a whole-genome sequence is generated for each. d, Phenotypic characterization and functional validation of metagenomics studies can be performed in vitro and in vivo.