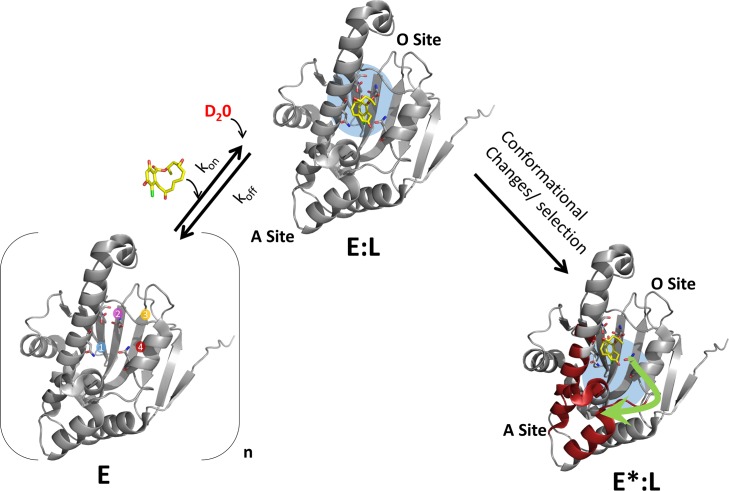
Fig 1. Mapping protein-ligand interactions by HDXMS.
Protein-ligand interactions can be analyzed by HDXMS by comparing deuterium exchange of the unliganded state of the protein with that bound to ligand (shown in yellow sticks). An ensemble view entails that the target protein (E) would exist in multiple conformations in the absence of ligand. Here a representative target protein is shown containing two sites- an orthosteric (O) site forming the ligand binding pocket (sites 1–4 are represented) and an allosteric (A) site. Deuterium exchange at the orthosteric site (O-site) (blue) is then governed by ligand binding kinetic parameters: kon, koff, concentration of ligand as well as the observed rate of HDX exchange, kex, which varies across different regions of the protein. The HDXMS output encompasses changes at the orthosteric O-site and long range conformational changes (red) at the allosteric A-site. Binding of ligand at the O-site (E:L) would result in decreased exchange while changes at the A-site (E*:L) could be reflected as decreases or increases in deuterium exchange.