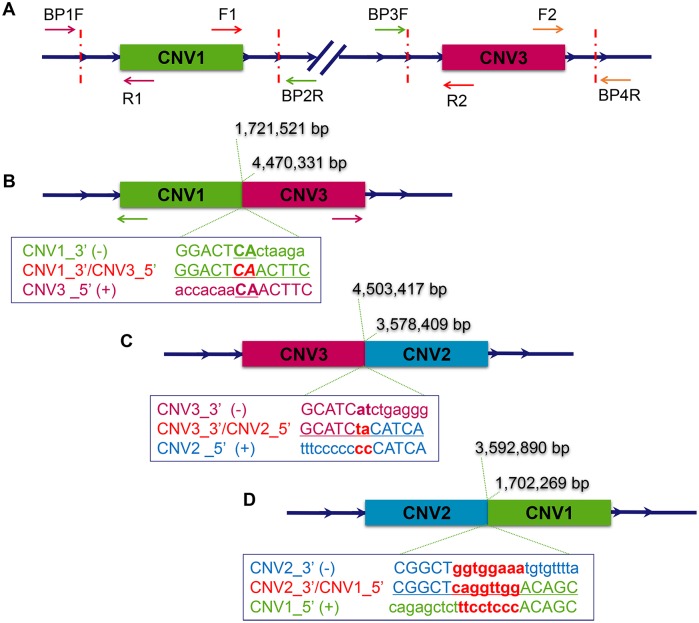
Fig 4. Analyzing the breakpoints of the copy number variations on GGA27 to clarify the rearrangement pattern.
The duplicated regions identified by array-CGH are illustrated by green (CNV1), blue (CNV2) and pink (CNV3) boxes, respectively. (A) The boundaries of the CNVs were tested using 8 primers indicated by the arrows located in the known duplicated region of CNV1 and CNV3. All possible amplifications were considered and performed in both Mb and wild-type chickens. (B) The breakpoints of both CNV1_3’ (1,721,521 bp) and CNV3_5’ (4,470,331 bp) were identified after sequencing the specifically-amplified PCR product obtained in Mb chickens using primer F1 and R2. An overlap of two nucleotides was detected in the junction region. Outward facing primers (green and pink arrows) were designed to analyze the other boundary of CNV1 and CNV3. (C) CNV2 was found to be located next to CNV3 in chickens with the Mb phenotype using a genome-walking strategy. The breakpoints of both CNV3_3’ (4,503,417 bp) and CNV2_5’ (3,578,409 bp) were verified by sequencing. A two-nucleotide insertion was found in the junction region. (D) The breakpoints of both CNV2_3’ (3,592,890 bp) and CNV1_5’ (1,702,269 bp) were confirmed through unmapped read alignment of whole genome re-sequencing data. An eight-base insertion was detected in the junction.