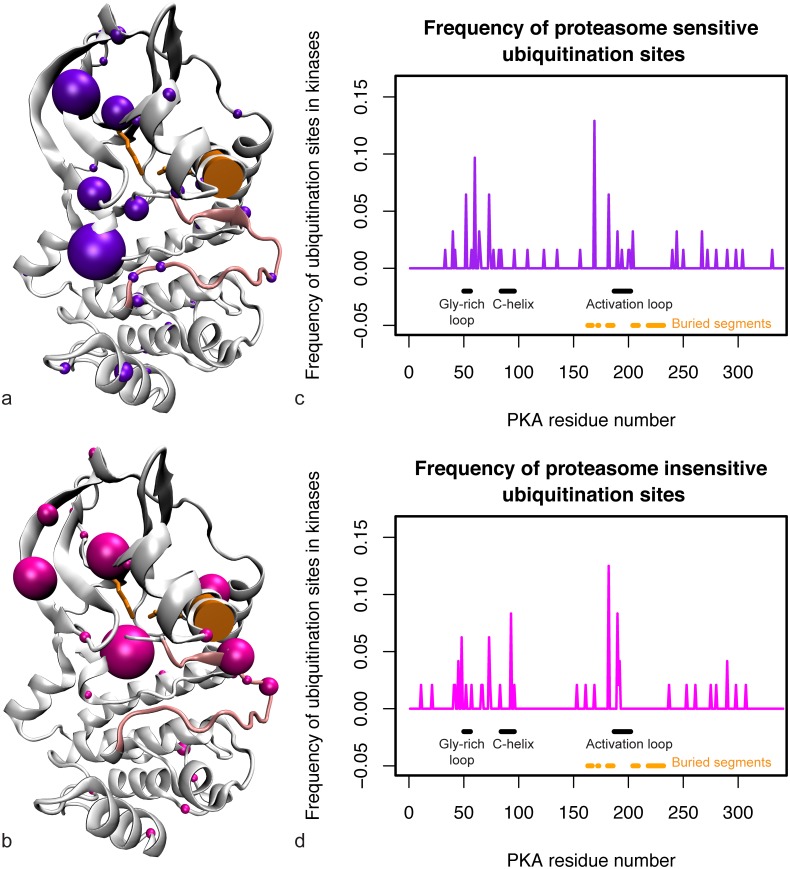
Fig 2. All proteasome-sensitive and proteasome-insensitive sites from human kinases.
(a) Proteasome-sensitive sites (purple spheres) mapped onto the PKA structure. Sphere size is proportional to the fraction of ubiquitin modifications that occur at that site. (b) Proteasome-insensitive sites (pink spheres) mapped onto the PKA structure. Sphere size is proportional to the fraction of ubiquitin modifications that occur at that site. The C-helix is highlighted by an orange cylinder, K369 on strand β3 of the N-lobe and E386 on the C-helix (forming the salt bridge that holds the C-helix in place) are shown with orange sticks, and the activation loop is shown in light pink. (c) Frequency of proteasome-sensitive ubiquitination sites on the kinase domain. (d) Frequency of proteasome-insensitive ubiquitination sites on the kinase domain. The ubiquitination sites across all kinases are mapped onto the PKA sequence on the x-axis, normalized by the total density on the kinase domain.