Figure 1. Molecular characteristics of patient-derived melanoma populations used in the study.
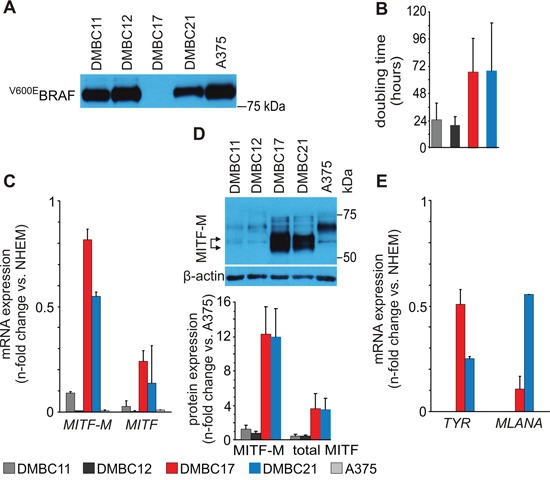
A. Western blot analysis of BRAF mutation status in patient-derived melanoma cell populations (DMBCs). An antibody recognizing BRAFV600E but not wild type BRAF was employed. A375, a melanoma cell line harboring BRAFV600E mutation was used as a positive control. B. Doubling time of melanoma cell populations assessed as metabolic activity of acid phosphatase. (n = 3) C. qRT-PCR analysis comparing basal levels of MITF-M and MITF. D. Western blot analysis comparing basal levels of MITF. A doublet of M isoform is indicated by arrows (top). MITF (total) and MITF-M protein levels were quantified relatively to their levels in A375 cells (bottom) (n = 3). E. qRT-PCR analysis comparing basal transcript levels of tyrosinase (TYR) and MLANA. Relative mRNA quantity of MITF and MITF-M (panel C), TYR and MLANA (panel E) is represented after normalization to RPS17 and the level in melanocytes (NHEM). As in DMBC11 and DMBC12 cells the expression of TYR and MLANA was several hundred fold lower than in NHEM, it is displayed as zero. DMBC, patient-derived melanoma populations obtained in Department of Molecular Biology of Cancer.
